Difference between revisions of "Team:Waterloo/Modeling/PAM Flexibility"
m |
(Added photo, needs to be formatted) |
||
| Line 9: | Line 9: | ||
<p>However, Kleinstiver et al. recently demonstrated modified spCas9 with altered PAM specificity. Cells were transformed with a plasmid bearing a toxic gene and an NGA adjacent protospacer, as well as a spCas9 gene with a randomly mutagenized PAM Identification Domain. Only cells capable of binding the NGA PAM sequence were able to cut the plasmid, and thus survive. By sequencing surviving clones, the authors were able to identify mutations in the PI domain that altered PAM sequence specificity. <cite ref="Kleinstiver2015"></cite></p> | <p>However, Kleinstiver et al. recently demonstrated modified spCas9 with altered PAM specificity. Cells were transformed with a plasmid bearing a toxic gene and an NGA adjacent protospacer, as well as a spCas9 gene with a randomly mutagenized PAM Identification Domain. Only cells capable of binding the NGA PAM sequence were able to cut the plasmid, and thus survive. By sequencing surviving clones, the authors were able to identify mutations in the PI domain that altered PAM sequence specificity. <cite ref="Kleinstiver2015"></cite></p> | ||
<p>Two mutants in particular showed both high specificity and activity for the NGA PAM sequence. The first mutant, referred to as VQR (D1135V,R1335Q,T1337R), worked equivalently on all four NGAN PAM sequences. The second mutant, EQR (D1135E, R1335Q, T1337R) was most specific to NGAG PAMs. A second round of testing also found a VRER variant ((D1135V,G1218R,R1335E,T1337R) that was specific to NGCG PAMs. <cite ref="Kleinstiver2015"></cite></p> | <p>Two mutants in particular showed both high specificity and activity for the NGA PAM sequence. The first mutant, referred to as VQR (D1135V,R1335Q,T1337R), worked equivalently on all four NGAN PAM sequences. The second mutant, EQR (D1135E, R1335Q, T1337R) was most specific to NGAG PAMs. A second round of testing also found a VRER variant ((D1135V,G1218R,R1335E,T1337R) that was specific to NGCG PAMs. <cite ref="Kleinstiver2015"></cite></p> | ||
| + | <figure> | ||
| + | <img src="/wiki/images/8/8c/Eqr_mutated_residues.png" alt="EQR Mutated Residues" style="width:500px;"/> | ||
| + | <figcaption>A PyMOL generated image of the three mutations found in the VQR spCas9 variant</figcaption> | ||
| + | <div class="img-att">i | ||
| + | <ul class="img-att-bubble"> | ||
| + | <li><a href="#">Link to original Photo</a></li> | ||
| + | </ul> | ||
| + | </div> | ||
| + | </figure> | ||
<p>These result indicate that spCas9 can display altered PAM specificity with relatively few mutants. Additionally, the data produced by Kleinstiver et al. also provided significant insight into the types of mutations that altered PAM specificity. This allows for a more targeted approach to engineering new spCas9 PAMs.</p> | <p>These result indicate that spCas9 can display altered PAM specificity with relatively few mutants. Additionally, the data produced by Kleinstiver et al. also provided significant insight into the types of mutations that altered PAM specificity. This allows for a more targeted approach to engineering new spCas9 PAMs.</p> | ||
<p>PAM Sites Differ between Natural Cas9 PRoteins</p> | <p>PAM Sites Differ between Natural Cas9 PRoteins</p> | ||
Revision as of 11:40, 18 September 2015
Modelling Engineered Cas9 PAM Flexibility
CRISPR-Cas9 is one of the most significant advances in genetic engineering in the last several years. However, one limitation of the S. pyogenes CRISPR-Cas9 (spCas9) is the NGG protospacer adjacent motif (PAM) required for binding to the target DNA site. While the 20bp protospacer can be easily specified with different sgRNAs, the PAM is essentially hardcoded into the Cas9 protein structure of the PAM-interacting (PI) domain. This limits the number of possible targets in a given genome, and thus the possible uses of spCas9 for genetic engineering. However, attempts to selectively modify Cas9 to bind new PAMs, specifically by changing two amino acids thought to bind directly to the PAM DNA sequence, failed.
However, Kleinstiver et al. recently demonstrated modified spCas9 with altered PAM specificity. Cells were transformed with a plasmid bearing a toxic gene and an NGA adjacent protospacer, as well as a spCas9 gene with a randomly mutagenized PAM Identification Domain. Only cells capable of binding the NGA PAM sequence were able to cut the plasmid, and thus survive. By sequencing surviving clones, the authors were able to identify mutations in the PI domain that altered PAM sequence specificity.
Two mutants in particular showed both high specificity and activity for the NGA PAM sequence. The first mutant, referred to as VQR (D1135V,R1335Q,T1337R), worked equivalently on all four NGAN PAM sequences. The second mutant, EQR (D1135E, R1335Q, T1337R) was most specific to NGAG PAMs. A second round of testing also found a VRER variant ((D1135V,G1218R,R1335E,T1337R) that was specific to NGCG PAMs.
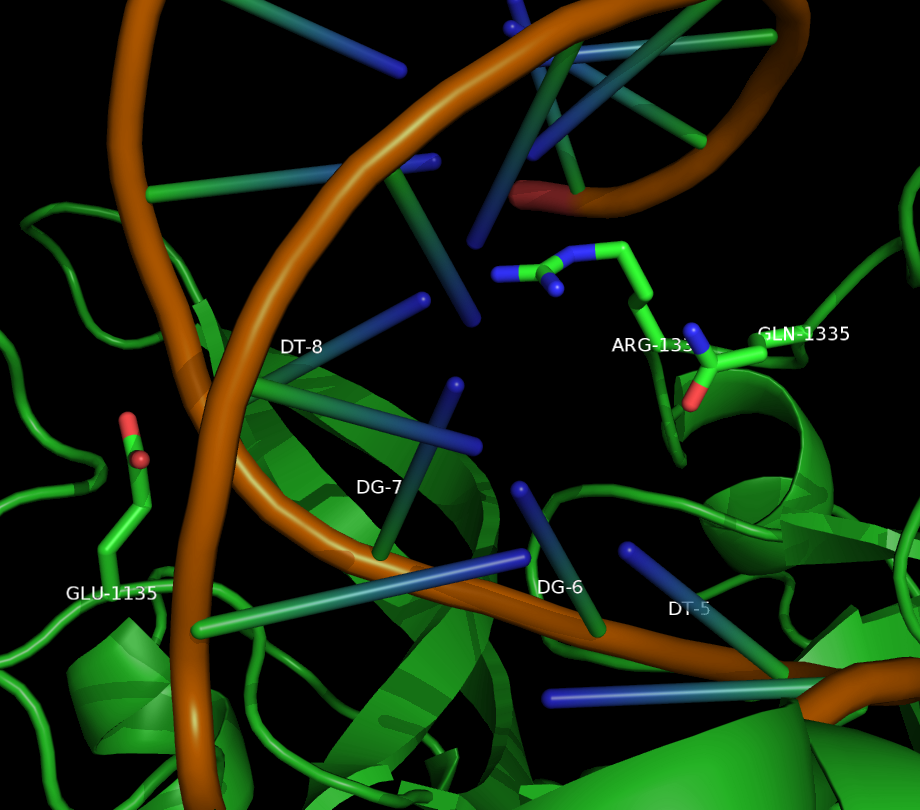
These result indicate that spCas9 can display altered PAM specificity with relatively few mutants. Additionally, the data produced by Kleinstiver et al. also provided significant insight into the types of mutations that altered PAM specificity. This allows for a more targeted approach to engineering new spCas9 PAMs.
PAM Sites Differ between Natural Cas9 PRoteins
Computational Engineering of PAM Flexibility
Modelling Binding with PyMOL and PyRosetta
As of June 2015, at least three different groups have produce experimental data on Cas9 structure. By using a catalytically dead version of spCas9 (dCas9), Anders et al. managed to determine the crystal structure of a Cas9 protein bound to both the sgRNA and the double stranded target DNA. The PDB file containing this structure can be found in the Protein Data Bank with ID 4UN3.
Canonical molecular dynamics data, mention source of PDB
Mutating PAM sites within PDB
Available PAM Affinity Data
Mention Klienstiver and
Engineering Pipeline
Summarize whole approach
Model Validation
Choice of DNA Mutation Software
Chimera VS 3DNA, give stats details
Patterns of Variation between Simulations
Simulations are Monte Carlo, we analyzed the differences between them
Wild-Type Cas9 PAM Affinities are Reproduced
Report Kendall's Tau, Pearson, Clustering Results
Mutant Cas9 PAM Affinities show Mixed Results
Mutating VQR/EQR, how EQR and VQR mutants change Cas9 visualizations, measures of distance from DNA bases to binding sites in the Cas9
Framework and Future Work
Proposed Tool
Link to software page re:pyrosetta
Future Work
Talk about adding other info like Gibb's Free Energy
Show a few graphs from Kleinstiver and link to dataset encouraging others to use