Difference between revisions of "Team:elan vital korea/Background"
| (13 intermediate revisions by 3 users not shown) | |||
| Line 61: | Line 61: | ||
background: url(https://static.igem.org/mediawiki/2015/9/9c/Lightbg_copy.jpg) 50% 0 no-repeat fixed; | background: url(https://static.igem.org/mediawiki/2015/9/9c/Lightbg_copy.jpg) 50% 0 no-repeat fixed; | ||
min-height:1000px; | min-height:1000px; | ||
| − | height: | + | height: auto; |
margin: 0 auto; | margin: 0 auto; | ||
width: 100%; | width: 100%; | ||
| Line 71: | Line 71: | ||
background: url(https://static.igem.org/mediawiki/2015/9/9c/Lightbg_copy.jpg) 50% 0 no-repeat fixed; | background: url(https://static.igem.org/mediawiki/2015/9/9c/Lightbg_copy.jpg) 50% 0 no-repeat fixed; | ||
height: 1000px; | height: 1000px; | ||
| − | + | height: auto; | |
margin: 0 auto; | margin: 0 auto; | ||
width: 100%; | width: 100%; | ||
| Line 81: | Line 81: | ||
#maintext2 { | #maintext2 { | ||
background: url(https://static.igem.org/mediawiki/2015/9/9c/Lightbg_copy.jpg) 50% 0 no-repeat fixed; | background: url(https://static.igem.org/mediawiki/2015/9/9c/Lightbg_copy.jpg) 50% 0 no-repeat fixed; | ||
| − | + | height: auto; | |
min-height:1000px; | min-height:1000px; | ||
margin: 0 auto; | margin: 0 auto; | ||
| Line 287: | Line 287: | ||
</font> | </font> | ||
</div> | </div> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<div class="inner"> | <div class="inner"> | ||
| Line 355: | Line 338: | ||
</div> | </div> | ||
<br> | <br> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<div class="inner"> | <div class="inner"> | ||
| Line 422: | Line 394: | ||
<P style="text-align:left;"> | <P style="text-align:left;"> | ||
<font color="black"> | <font color="black"> | ||
| − | |||
For example, the picture below represents the quorum sensing mechanism in the bacteria vibrio fisheri. Vibrio fisheri is a bacteria that produces bioluminescence, and is famous for revealing quorum sensing for the first time. Vibrio fisheri uses quorum sensing to produce light in high cell density, and researchers first discovered quorum sensing from examining vibrio fisheri.<br><br> | For example, the picture below represents the quorum sensing mechanism in the bacteria vibrio fisheri. Vibrio fisheri is a bacteria that produces bioluminescence, and is famous for revealing quorum sensing for the first time. Vibrio fisheri uses quorum sensing to produce light in high cell density, and researchers first discovered quorum sensing from examining vibrio fisheri.<br><br> | ||
| Line 432: | Line 403: | ||
</div> | </div> | ||
| − | <img class="displayed" src="https://static.igem.org/mediawiki/2015/ | + | <img class="displayed" src="https://static.igem.org/mediawiki/2015/3/3a/Graphics-25.png"> |
<br> | <br> | ||
| Line 468: | Line 439: | ||
<!-- Section #6 --> | <!-- Section #6 --> | ||
<section id="maintext2" data-speed="10" data-type="background"> | <section id="maintext2" data-speed="10" data-type="background"> | ||
| − | + | <br> | |
<div class="inner"> | <div class="inner"> | ||
<P style="text-align:left;"> | <P style="text-align:left;"> | ||
| Line 474: | Line 445: | ||
Quorum quenching takes advantage of the fact that quorum sensing also plays a role in expressing virulence, and interferes with the quorum sensing that produces virulence. There are many ways of utilizing quorum sensing for medicinal use, and one of the most intuitive and most well-known methods is quorum quenching. Quorum quenching takes advantage of the fact that quorum sensing also plays a role in expressing virulence, and interferes with the quorum sensing that produces virulence. However, for our project this year, we decided to focus on engineering a detection method for antibiotic resistance. For the project, we created a test plasmid and a reporter plasmid. We then transformed competent E. coli with the plasmids to produce a test cell and a reporter cell. As shown in the picture below, the test cell produces lactonase, which breaks down AHL, a common auto-inducer in gram-negative bacteria. And the reporter cell produces GFP (or luciferase) which creates a visible difference that we can detect. Both plasmids were engineered using the BioBrick DNA recombination process. With such a set up, it will be possible to detect the presence of the test cell, or lactonase.<br><br> | Quorum quenching takes advantage of the fact that quorum sensing also plays a role in expressing virulence, and interferes with the quorum sensing that produces virulence. There are many ways of utilizing quorum sensing for medicinal use, and one of the most intuitive and most well-known methods is quorum quenching. Quorum quenching takes advantage of the fact that quorum sensing also plays a role in expressing virulence, and interferes with the quorum sensing that produces virulence. However, for our project this year, we decided to focus on engineering a detection method for antibiotic resistance. For the project, we created a test plasmid and a reporter plasmid. We then transformed competent E. coli with the plasmids to produce a test cell and a reporter cell. As shown in the picture below, the test cell produces lactonase, which breaks down AHL, a common auto-inducer in gram-negative bacteria. And the reporter cell produces GFP (or luciferase) which creates a visible difference that we can detect. Both plasmids were engineered using the BioBrick DNA recombination process. With such a set up, it will be possible to detect the presence of the test cell, or lactonase.<br><br> | ||
| − | For the confirmation of our hypothesis, we conducted some experiments. Ideally, mixing AHL with the test cell will break down the AHL. And, adding the solution to reporter after that will not result in any fluorescence. But, if we do the same process with the control bacteria instead of the test cell, there will be fluorescence. As theorized, the control experiments produced fluorescence, but the experiments with the test cell produced no fluorescence. which means (no breakdown of signaling molecule should occur). | + | For the confirmation of our hypothesis, we conducted some experiments. Ideally, mixing AHL with the test cell will break down the AHL. And, adding the solution to reporter after that will not result in any fluorescence. But, if we do the same process with the control bacteria instead of the test cell, there will be fluorescence. As theorized, the control experiments produced fluorescence, but the experiments with the test cell produced no fluorescence. which means (no breakdown of signaling molecule should occur).<br> |
</font> | </font> | ||
</p> | </p> | ||
</div> | </div> | ||
| − | <img class="displayed" src="https://static.igem.org/mediawiki/2015/ | + | <img class="displayed" src="https://static.igem.org/mediawiki/2015/a/a0/Graphics-28.png" width="1000"> |
<br><br> | <br><br> | ||
| Line 544: | Line 515: | ||
<div class="inner"> | <div class="inner"> | ||
| − | <P style="text-align: | + | <P style="text-align:left;"> |
<font color="black"> | <font color="black"> | ||
Antibiotic Resistance Threat in the United States 2013, US Department of Health and Human Services, <br> | Antibiotic Resistance Threat in the United States 2013, US Department of Health and Human Services, <br> | ||
| Line 578: | Line 549: | ||
</div> | </div> | ||
| − | < | + | <div class="inner"> |
| − | + | <P style="text-align:left;"> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | <P style="text-align: | + | |
<font color="black"> | <font color="black"> | ||
Jean Livet, Tamily A. Weissman1, Hyuno Kang, Ryan W. Draft1, Ju Lu1, Robyn A. Bennis1, Joshua R. Sanes Jeff W. Lichtman,Transgenic strategies <br> | Jean Livet, Tamily A. Weissman1, Hyuno Kang, Ryan W. Draft1, Ju Lu1, Robyn A. Bennis1, Joshua R. Sanes Jeff W. Lichtman,Transgenic strategies <br> | ||
| Line 616: | Line 575: | ||
</font> | </font> | ||
</p> | </p> | ||
| − | + | </div> | |
<br> | <br> | ||
<br> | <br> | ||
Latest revision as of 14:50, 18 September 2015

BACKGROUND
Bacteria acquiring resistance to antibiotics pose serious health problem globally. Following last year’s example, the project of Elan Vital Korea for this year also is related to MRSA. This year, however, we have focused on early detection of MRSA infection using quorum sensing. Below, we have briefly described the health threats caused by MRSA, and have explained the quorum sensing method. Then, we have proceeded to the description of how we designed and implemented our experiments, and what results we have obtained. Finally, we have briefly outlined the implication of our results and future plans.

Threats of Antibiotics-Resistant Bacteria
Infection by antibiotic-resistant bacteria is a serious health threat worldwide including Korea and the United States of America. It is a serious threat primarily because, as the name suggests, bacteria have evolutionarily developed a resistance to antibiotics. It means, first of all, drugs don’t work. Furthermore, the spread of the antibiotic-resistant bacteria makes it more difficult to control or contain the spread of the infectious disease, because it undermines the effectiveness of treatment. And, it substantially increases the cost of healthcare, and the burden to society because it prolongs the treatment period and increases the likelihood of death. WHO declared that it “threatens the achievements of modern medicine” (Antimicrobial Resistance: Global Report on Surveillance 2014, WHO, 2014). Antimicrobial resistance already causes 700,000 deaths every year, which number is expected to 10 million annually by 2050 (An international legal framework to address antimicrobial resistance, WHO, 2015).
What makes the problem more pressing is that the data isbased on the reports of clinical samples from
laboratories, “predominantly in hospital settings” (Antimicrobial Resistance: Global Report on
Surveillance 2014, WHO, 2014, p. 70), which means community-acquired (compared to health-care associated) infections and uncomplicated infections are underrepresented.
Global Report on Surveillance 2014, WHO, 2014, p. 70), which means community-acquired (compared to health-care
associated) infections and uncomplicated infections are underrepresented.

Existing Methods Used for Detection
CDC’s efforts at outsmarting the antibiotic resistance focuses on 4 core actions: detect, respond, prevent and discover. The project is called AR Initiative (Detect and Protect Against Antibiotic
Resistance Initiative), which is an integral part of the CDC strategy to target investment aimed at AR.
Among the AR initiative, detection is the first step that impacts the whole controlling process.
Detecting antibiotic resistance quickly and effectively is crucial for determination of the treatment methods
for different patients as well as for quarantines to prevent it from becoming epidemic.
Currently, several methods are used for the detection of the antibiotic resistance. Most common and traditional
method is using growth inhibition assays performed in broth or by agar disc diffusion.
For clinically critical bacteria, diagnostic laboratories perform phenotypic-based analyses using standardized
susceptibility testing methods, usually in accordance with the guidelines published by the Clinical
and Laboratory Standards Institute.
Using the culture-based approach, it can take 1—2 days to produce results for fast-growing bacteria such as
Escherichia coli orSalmonella, but several weeks for slow-growing bacteria such as Mycobacterium tuberculosis.
Moreover, culturing only works for a small fraction of microbes; although most pathogens can be cultured
relatively easily thanks to years of accumulated experimental experiences, the vast majority of microbes cannot
grow outside their host environment, including pathogens such as Chlamydia orTrypanosomes.
Using newer molecular detection techniques for antibiotic resistance such as quantitative PCR (qPCR) or microarrays, we can determine the presence of specific resistance genes within hours, and we obtain improved diagnosis results. However, these culture-independent approaches target well-known and well-studied pathogens or resistance-causing genes only, and cannot be easily used for broader spectrum screening.
CDC dramatically innovated the detection process by adopting the Advanced Molecular Detection (AMD), which combines the latest pathogen identification technologies with bioinformatics and advanced epidemiology to more effectively understand, prevent and control infectious diseases. Using those technologies, it is possible to rapidly look for a microbe's match among thousands of reference samples in the microbe library. If no match is found, the whole genomic sequence of the microbe's DNA code can be taken, then quickly analyzed using disease detective works and bioinformatica to answer critical disease-response questions. However, this new method, while it sounds very interesting, is not to be completed until 2020, and still requires incubation, as well as being expensive.

Our Hypothesis: Possibility of Using Quorum
Sensing for Early Detection
Our team, Elan Vital Korea, addressed the problem of rapidly detecting antibiotic-resistant bacteria. We were interested in a rapid and efficient method of antibiotic resistance detection, and we believed that such a method could be engineered using quorum sensing. Our hypothesis was that we would be able to use quorum sensing – a method bacteria use to communicate with each other – to make the cells quickly report the existence of antibiotic-resistant bacteria
By quorum sensing, bacteria can perform many cooperative functions, such as biofilm formation, antibiotic production, motility, swarming, virulence, and much more. While most quorum sensing takes place between bacteria of the same species, there are cases of interspecies quorum sensing. Auto-inducers affect the gene expression of the bacteria once they reach a certain concentration threshold. Bacteria using quorum sensing usually produce small amounts of auto-inducers, so that the concentration of auto-inducers are affected by the concentration of the bacteria. In other words, quorum sensing, in essence, regulates gene expression in response to cell density. Using quorum sensing, bacteria are able to act in unison, as if they were a single organism.
Quorum sensing is widely used by various bacteria for various functions, so each uses a slightly different auto-inducer so the signals are not mixed up. In general, gram-negative bacteria use a class of molecules called N-acyl homoserine lactones, or AHL, while gram-positive bacteria use short processed polypeptides.
For example, the picture below represents the quorum sensing mechanism in the bacteria vibrio fisheri. Vibrio fisheri is a bacteria that produces bioluminescence, and is famous for revealing quorum sensing for the first time. Vibrio fisheri uses quorum sensing to produce light in high cell density, and researchers first discovered quorum sensing from examining vibrio fisheri.
In vibrio fishri, quorum sensing involves LuxI and LuxR as well as AHL. LuxI is the protein that produces AHL, and LuxR forms a complex with AHL to affect the regulation of genes. In this case, it produces luciferase, which produces bioluminescence. Furthermore, the process also boosts the production of LuxI, which creates a positive feedback loop. This AHL-LuxR quorum sensing mechanism is one of the most well known gram-negative quorum sensing pathways, and it can be engineered to affect almost any coding sequence we like.

For the project, we have developed a reporter cell that expresses GFP in the presence of the QS signaling molecule
acyl homoserine lactone (AHL). Our test cells (which act as a simulation of antibiotic-resistant bacteria) express lactonase, which breaks down AHL. In our experimental system, test cells should signify their presence by breaking down AHL and preventing GFP expression in reporter cells.

Experiment: Process and Results
There are many ways of utilizing quorum sensing for medicinal use, and one of the most intuitive and most well-known methods is quorum quenching.
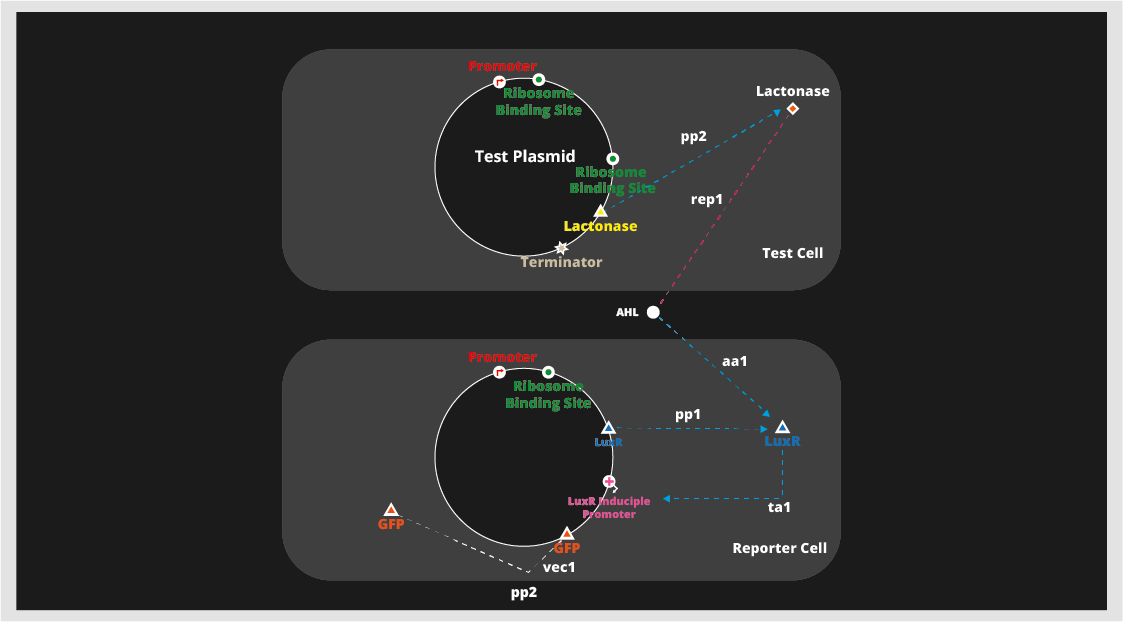
Quorum quenching takes advantage of the fact that quorum sensing also plays a role in expressing virulence, and interferes with the quorum sensing that produces virulence. There are many ways of utilizing quorum sensing for medicinal use, and one of the most intuitive and most well-known methods is quorum quenching. Quorum quenching takes advantage of the fact that quorum sensing also plays a role in expressing virulence, and interferes with the quorum sensing that produces virulence. However, for our project this year, we decided to focus on engineering a detection method for antibiotic resistance. For the project, we created a test plasmid and a reporter plasmid. We then transformed competent E. coli with the plasmids to produce a test cell and a reporter cell. As shown in the picture below, the test cell produces lactonase, which breaks down AHL, a common auto-inducer in gram-negative bacteria. And the reporter cell produces GFP (or luciferase) which creates a visible difference that we can detect. Both plasmids were engineered using the BioBrick DNA recombination process. With such a set up, it will be possible to detect the presence of the test cell, or lactonase.
For the confirmation of our hypothesis, we conducted some experiments. Ideally, mixing AHL with the test cell will break down the AHL. And, adding the solution to reporter after that will not result in any fluorescence. But, if we do the same process with the control bacteria instead of the test cell, there will be fluorescence. As theorized, the control experiments produced fluorescence, but the experiments with the test cell produced no fluorescence. which means (no breakdown of signaling molecule should occur).
Expected Benefits
Thanks to bacteria’s ability to make quick and profound changes in gene transcription, quorum sensing can be
used to detect a low amount of signaling molecules and report their presence quickly. With further
research and thorough engineering applications, it may be possible to detect other antibiotic-resistant bacteria that are unknown until now.
If it is proven as valid and effective through sufficient tests, this technique could be disseminated to
hospitals and clinics to test the presence of antibiotic-resistant bacteria. We hope that this technique, if properly adjusted for functional advancement, can detect antibiotic-resistant bacteria in a relatively short time with only a small amount of sample secured from the patient. This would provide an advantage over the traditional detection methods, culture-based approaches which require one or several days of incubation period.
Because chemicals involved in species-specific quorum sensing is very specific, it might be possible to
dramatically resolve the problem of overnight incubation. Because an initial sample from
a patient is usually contaminated and has only a small concentration of the wanted bacteria, it is often
impossible to detect any antibiotic-resistance without purification and amplification through overnight incubation. But because species-specific quorum sensing involves biochemical that are highly specific, and the quorum sensing chemicals are not affected as much by the contamination, the method utilizing quorum sensing might be applied with relatively less purification processes. Also, because
some quorum sensing mechanisms have built in positive feedback, with the right engineering, the mechanism could work with only a little amplification process.
More innovative detection methods such as quantitative PCR(qPCR) or microarrays, and advanced molecular
detection (AMD) are based on accumulated previous data and, thus, render very accurate results, but
they require complicated procedures and heavy equipment. On the other hand, this quorum sensing-based detection
method will provide benefits to patients with handy procedure and quicker detection results. We believe quicker
and easy detection of antibiotic-resistant bacteria will lead to better containment of such dangerous bacterial strains.
Reference
Antibiotic Resistance Threat in the United States 2013, US Department of Health and Human Services,
Center for Disease Control and Prevention About Quorum Sensing
Annual Review of Microbiology, Volume 55:pp 165-199 (volume publication date, October 2001) Melissa B.Miller and Bonnie L.
Bassler Department of Molecular Biology, Princeton University, Princeton, New Jersey
Bacterial Quorum Sensing: Its Role in Virulence and Possibilities for Its Control Steven T. Rutherfold and Bonnie L.Bassler.
Cold Spring Harb Perspect Med. 2012.2, Cold Spring Harbor Laboratory Press
Quorum Sensing: Bacteria Talk Sense Costi D. Sifri, Oxford Journals, Volume 47, Issue 8 Pp 1070-1076, 2015
Infectious Diseases Society of America
Bacterial Quorum Sensing in Pathogenic Relationships Teresa R. de Kievit, Barbara H.Iglewski, Infection and Immunity,
Volume 68, September 2000, 2000 American Society for Microbiology
Emily R. M. Sydnor and Trish M. Perl, Epidemiology and Infection Control in Acute-Care Settings, , Clinical Microbiology Reviews, 2011, January
Valdivia RH, Cormack BP. 2005. The uses of green fluorescent protein in prokaryotes. Methods Biochem. Anal.47:163–178 [PubMed]
Dubnau D. 1991. Genetic competence in Bacillus subtilis. Microbiol. Rev.55:395–424 [PMC free article][PubMed]
Errington J. 2003. Regulation of endospore formation in Bacillus subtilis. Nat. Rev. Microbiol.1:117–126 [PubMed]
Kearns DB, Losick R. 2005. Cell population heterogeneity during growth of Bacillus subtilis. Genes Dev.19:3083–3094 [PMC free article][PubMed]
NATIONAL ACTION PLAN TO PREVENT HEALTH CARE-ASSOCIATED INFECTIONS: ROAD MAP TO ELIMINATION APRIL 2013
FACT SHEET: Obama Administration Releases National Action Plan to Combat Antibiotic-Resistant Bacteria, March 27, 2015
Jean Livet, Tamily A. Weissman1, Hyuno Kang, Ryan W. Draft1, Ju Lu1, Robyn A. Bennis1, Joshua R. Sanes Jeff W. Lichtman,Transgenic strategies
for combinatorial expression of fluorescent proteins in the nervous system, Nature Volu 450, November 2007
Peter Novick and Randy Schekman,Secretion and cell-surface growth are blocked in a temperaturesensitive mutant of Saccharomyces cerevisiae,
Cell Biology, Vol. 76, No. 4, pp. 1858-1862, April 1979Department of Biochemistry, University of California, Berkeley,
Stewart, P.S., J. Rayner, F. Roe, and W.M. Rees, Biofilm Penetration and Disinfection Efficacy of Alkaline Hypochlorite and
Chlorosulfamates Jourmal of Applied Micro biology (2001) 91(3), 525-532.
de Nys, R., Steinberg, P.D., Willemsen, P., Dworjanyn, S.A., Gabelish, C.L., and King, R.J, Broad spectrum effects of
secondary metabolites from the red alga Delisea Pulchra in antifouling assays, Biofouling, (1995),
Hasting, J.W. and Greenberg, E.P., Quorum sensing: the explanation of a curious phenomenon reveals a common characteristic
of bacteria, Journal of Bacteriology,(1999) 181(9), 2667-2668.
Intercellular communication and quorum sensing in microorganism, Science Progress, (1998) 81(1), 69-80.
Gray, K.M. Intercellular communication and group behavior in bacteria, Trends in Microbiology, (1997) 5(5), 184-188.
Withers, H. Swift, S., Williams, P. Quorum sensing as an integral component of gene regulatory networks in Gram- negative bacteria,
Current Opinion in Microbiology, (2001), 4, 186-193.


