Difference between revisions of "Team:DTU-Denmark/Project/Intein"
(Created page with "<html> <head> <meta name="viewport" content="width=device-width, initial-scale=1, maximum-scale=1, user-scalable=no"> <meta http-equiv="X-UA-Compatible" content="IE=...") |
(No difference)
|
Revision as of 16:51, 18 September 2015
Intein
During our experimental work in the laboratory, we encountered many obstacles, when we tried to heterologously express the 40.8kb tyrocidine operon in Bacillus subtilis [read more link MISSING]. This inspired us to think of alternative approaches to produce of nonribosomal peptide drugs than by nonribosomal peptide synthetases (NRPS). We came up with a peculiar idea of generating short cyclised peptides similar in length to tyrocidine using self-splicing proteins. Inteins are short self-splicing proteins that have no functional role in proteins other than catalyzing their own excision after translation. This forms a peptide bond between the two adjent amino acids of the inteins (Figure #MISSING). If the N-intein and C-intein are placed at each end of a polypeptide; cyclisation would give a cyclised protein - or in our case a short cyclic peptide.
Achivements
- Designed construct for expression of two short cyclic peptides using BioBrick BBa_K1362000 and two short primers.
- Made the cyclisation construct in the laboratory.
- Analyzed constructs, though no products could be detected.
- Improved characterization of BioBrick BBa_K1362000.
Background
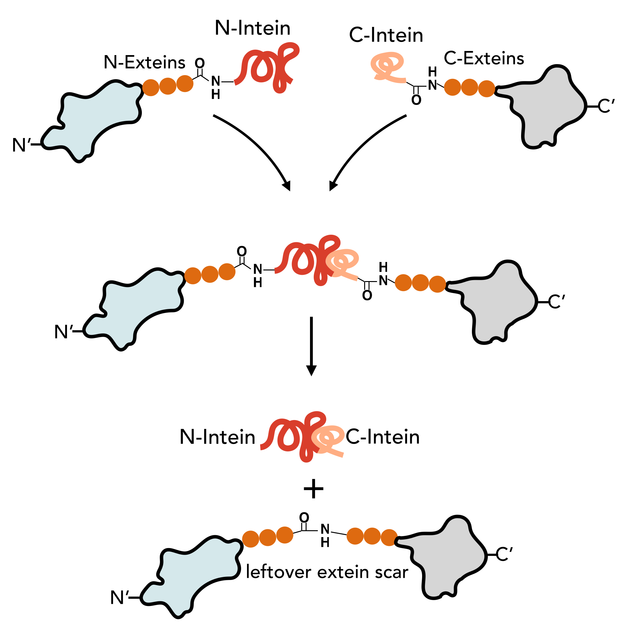 |
| Figure #Missing N-intein and C-intein is spliced out by forming a leftover extein scar of the size of the N-extein and C-extein. The size and amino acid sequenceof the scar depends |
Nonribosomal peptide synthetases do not have monopolly on production of bioactive nonribosomal peptides. CyBase contains 818 entries of ribosomal head-to-tail cyclic peptides from 105 different species [1]. They varry in length from 8 to 80 amino acids may contain internal cysteine bridges that stabilises the peptide [2]. Together natural cyclic peptides spans a huge diversity and due to their high thermostability and reported activities such as anti-HIV, anti-cancer, and anti-bacterial, cyclisation of peptides may be used to increase bioavailability of peptide drugs. Alternatively, as we hypothesied, their biosynthesis may be the key to synthese nonribosomal peptide drugs, independently of NRPS.
Cyclisation of these protein occurs post translation and is mediated by short peptides that have autocatalytic activity. In one mechanism (Figure #MISSING), self-splicing peptide sequences (inteins) bind and catalyze their own excission while forming a peptide bond between the N-extein and C-extein. Numerous of inteins have been characterized including split intein Npu DnaE from Nostoc punctiforme PCC73102 (Npu) (Figure #MISSING) [3,4]. When the Npu DnaE intein auto-catalyze its own excisions, it leaves the six amino acid extein-scar behind. The native scar (AEY-CFN) can be modified at some positions and still yield splicing [4]. The cysteine of the first position in the C-extein was the only amino acid that could not be substituted.
Experimental Design
The BioBrick BBa_K1362000 was introduced by Heidelberg in 2014. It contains Npu DnaE inteins surrounding a RFP selection marker, which can be cut out with restriction enzyme BsaI. Digestion of BBa_K1362000 with BsaI generates to different overhangs, thereby making it possible to insert the coding sequence (CDS) for protein of interest. The BioBrick was used to cyclise different proteins, but it was not used to cyclise short peptides.
We decided to try and cyclise an synthetic ten amino acid peptide sequence, which was similar to tyrocidine. In addition, we searched the cyclic peptide database for bioactive cyclic peptide which contained part of the extein scar. The sequences for both native peptides along with the sequence chosen for cyclisation experiments are shown in Table #MISSING.
| Table MISSING Peptides selected for this study. Npu DnaE requires a cystine at position 1 in the extein [4]. For tyrocidine the native extein sequence was kept. | ||||||||||||
|
The peptide insert was easily created by designing two complementary oligos that will generate overhangs compatible with ligation into BBa_K1362000 (TABLE #MISSING):
| Table #MISSING Oligos ordered for cycloviolacin Y1 and tyrocidine-like (see Table #MISSING). Oligos were annealed by heating 10µM of the paired oligos to 94° C in a thermocycler. After 2 minutes the thermocycler was turned off to allow the oligos to anneal while cooling down. | ||||
|
Results
MISSING
Transformants for analysis were re-streaked onto LB agar plates containing 6
It was not possible to detect cyclised proteins from cell-extracts my MALDI-TOF with a total of twelwe samples; six from each cyclic peptide. The size of the unspliced protein is 19.4 kDA (cycloviolacin Y1) and 17.2 kDa (tyrocidine-like), but it was not possible to detect bands at this molecular weight by SDS-PAGE (data not shown). Due to time constraints, the constructs were not sequenced.
Discussion
It was not possible to produce short cyclic peptides using the Npu DnaE. It was not further investigated, if it was due to the short length or the modification of the extein/intein sequence for cycloviolacin Y1. It may be impossible for the intein to cyclize the construct due to their short sequence. Despite being unsuccesfull, it has been reported that cyclotides similar to cyclovilacin Y1 could be cyclised by Npu DnaE in E. coli [5]. It has however to our knowledge not been able to use Npu DnaE to succesfully cyclise a ten amino acid peptide. It may be possible to achieve cyclisation of these peptides by exploring the diversity of inteins from different organisms.
References
- CyBase. September 15, 2015. http://www.cybase.org.au
- Thorstholm L, Craik DJ. Discovery and applications of naturally occurring cyclic peptides. Drug Discov Today Technol. 2012;9:e13–e21. doi: 10.1016/j.ddtec.2011.07.005.
- Zettler, J., Schütz, V., & Mootz, H. D. (2009). The naturally split Npu DnaE intein exhibits an extraordinarily high rate in the protein trans-splicing reaction. FEBS Letters, 583(5), 909–914. doi:10.1016/j.febslet.2009.02.003
- Cheriyan, M., Pedamallu, C. S., Tori, K., & Perler, F. (2013). Faster Protein Splicing with the Nostoc punctiforme DnaE Intein Using Non-native Extein Residues. Journal of Biological Chemistry, 288(9), 6202–6211. doi:10.1074/jbc.m112.433094
- Jagadish, K., Borra, R., Lacey, V., Majumder, S., Shekhtman, A., Wang, L., Camarero, J.A. Expression of fluorescent cyclotides using protein trans-splicing for easy monitoring of cyclotide-protein interactions. Angew Chem Int Ed Engl. 2013 March 11; 52(11): 3126–3131.
Department of Systems Biology
Søltofts Plads 221
2800 Kgs. Lyngby
Denmark
P: +45 45 25 25 25
M: dtu-igem-2015@googlegroups.com