Difference between revisions of "Team:HUST-China/Description"
WangjieLiu (Talk | contribs) |
|||
| (67 intermediate revisions by 8 users not shown) | |||
| Line 6: | Line 6: | ||
<title>Team:HUST-China:Description</title> | <title>Team:HUST-China:Description</title> | ||
| − | <link href=" | + | <link rel="stylesheet" type="text/css" href="https://2015.igem.org/Team:HUST-China/CSS?action=raw&ctype=text/css" /> |
| − | <script src=" | + | |
| − | <script src=" | + | <script type="text/javascript" src="https://2015.igem.org/Team:HUST-China/jquery?action=raw&ctype=text/javascript"></script> |
| + | |||
| + | <script type="text/javascript" src="https://2015.igem.org/Team:HUST-China/JS?action=raw&ctype=text/javascript"></script> | ||
<style> | <style> | ||
| Line 45: | Line 47: | ||
top: 0px;} | top: 0px;} | ||
| − | + | table th,td{text-align:center;} | |
.picture{ width: 750px; | .picture{ width: 750px; | ||
height: auto; | height: auto; | ||
| Line 78: | Line 80: | ||
z-index:1;} | z-index:1;} | ||
| − | + | .first-menu{float: left; | |
| − | + | ||
font-size:19px; | font-size:19px; | ||
| − | max-width: | + | max-width: 20%; |
| − | padding:5px;} | + | padding-top:5px;} |
.other-menu{float: left; | .other-menu{float: left; | ||
| − | margin-left:1 | + | margin-left:1%; |
font-size:19px; | font-size:19px; | ||
| − | max-width: | + | max-width: 20%; |
| − | padding:5px;} | + | padding-top:5px;} |
| + | |||
.navbar .nav > li .dropdown-menu {margin: 0;} | .navbar .nav > li .dropdown-menu {margin: 0;} | ||
| Line 115: | Line 117: | ||
| − | #pic{ background-image: url(https://static.igem.org/mediawiki/2015/ | + | #pic{ background-image: url(https://static.igem.org/mediawiki/2015/5/52/HUST_PROJECT_6.JPG |
| + | |||
| + | ); | ||
width:1349px; | width:1349px; | ||
height:650px; | height:650px; | ||
| Line 131: | Line 135: | ||
} | } | ||
.pic_a{float: left; | .pic_a{float: left; | ||
| − | margin-top: | + | margin-top:30px; |
width: 1349px; | width: 1349px; | ||
text-align:center; | text-align:center; | ||
| Line 163: | Line 167: | ||
text-align: left; | text-align: left; | ||
float: left; | float: left; | ||
| − | font-size:16px;} | + | font-size:16px; |
| + | width: 850px} | ||
.description p{font-size: 16px; | .description p{font-size: 16px; | ||
width:809px;} | width:809px;} | ||
| Line 203: | Line 208: | ||
function showdes(){ | function showdes(){ | ||
$('#to_des').click(function(){ | $('#to_des').click(function(){ | ||
| − | $("html,body").animate({scrollTop: | + | $("html,body").animate({scrollTop:650},800); |
}); | }); | ||
} | } | ||
| Line 217: | Line 222: | ||
<!--悬浮菜单--> | <!--悬浮菜单--> | ||
| + | <!--悬浮菜单--> | ||
<div class="navbar"> | <div class="navbar"> | ||
<div class="navbar-inner"> | <div class="navbar-inner"> | ||
| Line 223: | Line 229: | ||
<li class="first-menu"><a href="https://2015.igem.org/Team:HUST-China">HOME</a> | <li class="first-menu"><a href="https://2015.igem.org/Team:HUST-China">HOME</a> | ||
</li> | </li> | ||
| − | <li class="dropdown other-menu" id="accountmenu"><a href="https://2015.igem.org/Team:HUST-China/ | + | <li class="dropdown other-menu" id="accountmenu"><a href="https://2015.igem.org/Team:HUST-China/Project">PROJECT<b class="caret"></b></a> |
<ul class="dropdown-menu"> | <ul class="dropdown-menu"> | ||
<li><a href="https://2015.igem.org/Team:HUST-China/Background">Background</a></li> | <li><a href="https://2015.igem.org/Team:HUST-China/Background">Background</a></li> | ||
| Line 233: | Line 239: | ||
</li> | </li> | ||
| − | <li class="dropdown other-menu" id="accountmenu"><a href="https://2015.igem.org/Team:HUST-China/ | + | <li class="dropdown other-menu" id="accountmenu"><a href="https://2015.igem.org/Team:HUST-China/Wetlab">WETLAB<b class="caret"></b></a> |
<ul class="dropdown-menu"> | <ul class="dropdown-menu"> | ||
<li><a href="https://2015.igem.org/Team:HUST-China/Results">Results</a></li> | <li><a href="https://2015.igem.org/Team:HUST-China/Results">Results</a></li> | ||
| Line 245: | Line 251: | ||
</li> | </li> | ||
| − | <li class="dropdown other-menu" id="accountmenu"><a href="https://2015.igem.org/Team:HUST-China/ | + | <li class="dropdown other-menu" id="accountmenu"><a href="https://2015.igem.org/Team:HUST-China/Parts">PARTS<b class="caret"></b></a> |
<ul class="dropdown-menu"> | <ul class="dropdown-menu"> | ||
<li><a href="https://2015.igem.org/Team:HUST-China/Basic_part">Basic Parts</a></li> | <li><a href="https://2015.igem.org/Team:HUST-China/Basic_part">Basic Parts</a></li> | ||
<li class="divider"></li> | <li class="divider"></li> | ||
| − | <li><a href="https://2015.igem.org/Team:HUST-China/ | + | <li><a href="https://2015.igem.org/Team:HUST-China/Basic_part#2">Composite Parts</a></li> <li class="divider"></li> |
| + | <li><a href="https://2015.igem.org/Team:HUST-China/Part_Collection">Part Collection</a></li> | ||
</ul> | </ul> | ||
</li> | </li> | ||
| Line 255: | Line 262: | ||
<li class="dropdown other-menu" id="accountmenu"><a href="https://2015.igem.org/Team:HUST-China/Modeling">MODELING<b class="caret"></b></a> | <li class="dropdown other-menu" id="accountmenu"><a href="https://2015.igem.org/Team:HUST-China/Modeling">MODELING<b class="caret"></b></a> | ||
<ul class="dropdown-menu"> | <ul class="dropdown-menu"> | ||
| + | <li><a href="https://2015.igem.org/Team:HUST-China/Modeling">Overiew</a></li> | ||
| + | <li class="divider"></li> | ||
<li><a href="https://2015.igem.org/Team:HUST-China/Modeling on Cellular Level">Modeling on Cellular Level</a></li> | <li><a href="https://2015.igem.org/Team:HUST-China/Modeling on Cellular Level">Modeling on Cellular Level</a></li> | ||
<li class="divider"></li> | <li class="divider"></li> | ||
| Line 262: | Line 271: | ||
| − | + | <li class="dropdown other-menu" id="accountmenu"><a href="https://2015.igem.org/Team:HUST-China/Practices">HUMAN PRACTICES</a> | |
| − | <li class="dropdown other-menu" id="accountmenu"> | + | |
| − | + | ||
| − | + | ||
</li> | </li> | ||
| − | <li class="dropdown other-menu" id="accountmenu"> | + | <li class="dropdown other-menu" id="accountmenu"><a href="https://2015.igem.org/Team:HUST-China/Safety">OTHERS<b class="caret"></b></a> |
| − | + | ||
<ul class="dropdown-menu"> | <ul class="dropdown-menu"> | ||
<li><a href="https://2015.igem.org/Team:HUST-China/Safety">Safety</a></li> | <li><a href="https://2015.igem.org/Team:HUST-China/Safety">Safety</a></li> | ||
| Line 292: | Line 298: | ||
<!--标题栏--> | <!--标题栏--> | ||
<div id="pic" > | <div id="pic" > | ||
| − | <img class="title" src="https://static.igem.org/mediawiki/2015/ | + | <img class="title" src="https://static.igem.org/mediawiki/2015/2/2d/HUST_PROJECT_3.PNG"/> |
<br> | <br> | ||
<div class="pic_a" > | <div class="pic_a" > | ||
| − | < | + | <h3 align="center" style="color:white"><b>click it~</b></h3> |
<img style="cursor:pointer;" id="to_des" src="https://static.igem.org/mediawiki/2015/8/80/White.png"/> | <img style="cursor:pointer;" id="to_des" src="https://static.igem.org/mediawiki/2015/8/80/White.png"/> | ||
</div> | </div> | ||
| Line 308: | Line 314: | ||
<li class="li"><a href="#supporting" class="btn btn-default btn-lg">Supporting system</a></li> | <li class="li"><a href="#supporting" class="btn btn-default btn-lg">Supporting system</a></li> | ||
<li class="li"><a href="#flocculating" class="btn btn-default btn-lg">Flocculating system</a></li> | <li class="li"><a href="#flocculating" class="btn btn-default btn-lg">Flocculating system</a></li> | ||
| − | <li class="li"><a href="#host" class="btn btn-default btn-lg"> | + | <li class="li"><a href="#host" class="btn btn-default btn-lg">Yarrowia lipolytica</a></li> |
| − | <li class="li"><a href="#improvement" class="btn btn-default btn-lg">Improvement</a></li> | + | <li class="li"><a href="#improvement" class="btn btn-default btn-lg">Biobrick Improvement</a></li> |
<li class="li"><a href="#reference" class="btn btn-default btn-lg">References</a></li> | <li class="li"><a href="#reference" class="btn btn-default btn-lg">References</a></li> | ||
</ul> | </ul> | ||
| Line 315: | Line 321: | ||
<!--描述--> | <!--描述--> | ||
| − | + | <video width="800" height="400" controls="controls" style="margin-top:50px;margin-left:300px;z-index:-2"> | |
| − | + | <source src="https://static.igem.org/mediawiki/2015/8/89/IGEMHUSTVIDEO.mp4" type="video/webm"> | |
| − | + | Your browser does not support the video tag. | |
| + | </video> | ||
| + | <div class="description" style="margin-top:0px;margin-bottom:30px"> | ||
| + | <br><br> | ||
| + | <p>According to requirement of iGEM wiki construction, we hosted our cartoon instruction on iGEM server. For those can not play this video on your Browser such as IE11, please turn to view our wiki with google chrome or directly donwload this video <b><a href="https://static.igem.org/mediawiki/2015/8/89/IGEMHUSTVIDEO.mp4">HERE</a></b> from iGEM server.</p> | ||
| + | </div> | ||
| + | <div align="center" class="description"><a name="description"></a><br> | ||
<div class="dongxi"></div> | <div class="dongxi"></div> | ||
<h2 style="color:black" align="left"><b>1.Description</b></h2> | <h2 style="color:black" align="left"><b>1.Description</b></h2> | ||
| Line 324: | Line 336: | ||
<div > | <div > | ||
<img class="picture" src="https://static.igem.org/mediawiki/2015/8/8a/Hust_Figure1.png"> | <img class="picture" src="https://static.igem.org/mediawiki/2015/8/8a/Hust_Figure1.png"> | ||
| − | <div class="pizhu">Figure | + | <div class="pizhu">Figure 1:The complete circuit</div> |
</div> | </div> | ||
| − | <p>As mentioned above, | + | <p>As mentioned above, our design is briefly divided into three parts: Darkness induction system, flocculating system and supporting system. The latter two are both under control of the former one with the same upstream promoter Anb1. Fusion protein CRY2-BD and CIB1-AD are respectively expressed by constitutive promoter pTEF and pADH1. CRY2-BD has high affinity to bind CIB1-AD only when induced by blue light (even contained in nature sunlight) . This interaction can activate pGal1 owing to the combination between CIB1 and CRY2’s special structures thus induce the downstream promoter pGal1 and express ROX1 gene. Then expressed ROX1 protein can inhibit the promoter Anb1. So our engineered circuit is turned off when exposed to light. On the contrary, in darkness, the structural-reversed CRY2 will dissociate with CIB1. Without CRY2-BD’s target function, AD is hard to combine and active pGal1 in the complex environment of nucleus, so the downstream ROX1 is no longer transcribed. With the degradation of translated ROX1 protein, pAnb1 starts to express the downstream gene as the supporting matter and flocculating agent :LIP2 pro-Si-tag-YLcwp3 and LIP2-pro-Mcfp-3 fusion proteins(shown in Fig 1). </p> |
| − | + | </div> | |
| − | + | <div class="description"><a name="darkness"></a><br> | |
<div class="dongxi"></div> | <div class="dongxi"></div> | ||
| − | <h2 style="color:black" align="left"><b>2.Darkness | + | <h2 style="color:black" align="left"><b>2.Darkness Induction System</b></h2> |
| − | + | ||
| − | <img | + | <div> |
| − | + | <img style="width:400px;height:250px;float:left" class="picture" src="https://static.igem.org/mediawiki/2015/6/6a/Hust_figure2.png"> | |
| − | + | <img style="width:400px;height:250px;float:left" class="picture" src="https://static.igem.org/mediawiki/2015/e/e1/Hust_1.png"> | |
| − | + | <div class="pizhu">Figure 2:Darkness induction system</div> | |
| − | + | </div> | |
| − | + | ||
| − | <div class="pizhu">Figure | + | |
| − | + | ||
<p>The first one is the expression control system. Each system should apply a kind of trigger of working system. While a number of inducible systems are available for controlling protein expression in yeast, almost none of them make sense for us, as our project’s practical applications are currently considered to exist mainly in marine environment, which makes the amount of chemical inducers to be used a huge burden.</p> | <p>The first one is the expression control system. Each system should apply a kind of trigger of working system. While a number of inducible systems are available for controlling protein expression in yeast, almost none of them make sense for us, as our project’s practical applications are currently considered to exist mainly in marine environment, which makes the amount of chemical inducers to be used a huge burden.</p> | ||
| − | <p> So we give up several common promoters including those naturally responsive to molecules such as galactose, methionine, and copper, or engineered regulatory systems that respond to orthogonal molecules such as estrogen (Gal-ER-VP16)<sup>[1]</sup> or doxycycline (Tet-OFF)<sup>[2]</sup> .In fact, some of them are really robust and well designed pathways. However, we find a better method instead, a darkness induction system based on light control yeast two-hybrid principles, because darkness is the most common environment factor that our Euk.cement cells will face in working.</p> | + | <p> So we give up several common promoters including those naturally responsive to molecules such as galactose, methionine, and copper, or engineered regulatory systems that respond to orthogonal molecules such as estrogen (Gal-ER-VP16)<sup>[1]</sup> or doxycycline (Tet-OFF)<sup>[2]</sup> .In fact, some of them are really robust and well-designed pathways. However, we find a better method instead, a darkness induction system based on light control yeast two-hybrid principles, because darkness is the most common environment factor that our Euk.cement cells will face in working.</p> |
<p>Our original idea comes from dimers that allow inducible control of protein-protein interactions, which are powerful tools for manipulating biological processes. On this basis, we successfully utilized a split transcription factor that is reconstituted by light-dependent protein–protein interactions as in yeast two-hybrid systems. </p> | <p>Our original idea comes from dimers that allow inducible control of protein-protein interactions, which are powerful tools for manipulating biological processes. On this basis, we successfully utilized a split transcription factor that is reconstituted by light-dependent protein–protein interactions as in yeast two-hybrid systems. </p> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<p> Compared to those chemical-genetic tools, the light control system not only meets our demand, but also possesses many other advantages. Despite the general success of chemical inducers, the potential problems, such as toxic, nonspecific, or pleiotropic effects of these treatments can impose limitations on their use in sensitive chassis or environment. Also, light control is far more fast, precise, low cost and easy. It even allows exquisite dose-dependent control over protein levels, as light can be delivered for defined amounts of time and be instantly removed.</p> | <p> Compared to those chemical-genetic tools, the light control system not only meets our demand, but also possesses many other advantages. Despite the general success of chemical inducers, the potential problems, such as toxic, nonspecific, or pleiotropic effects of these treatments can impose limitations on their use in sensitive chassis or environment. Also, light control is far more fast, precise, low cost and easy. It even allows exquisite dose-dependent control over protein levels, as light can be delivered for defined amounts of time and be instantly removed.</p> | ||
| Line 358: | Line 362: | ||
<div> | <div> | ||
<img class="picture" src="https://static.igem.org/mediawiki/2015/f/fe/HUST_yl2.0.png"> | <img class="picture" src="https://static.igem.org/mediawiki/2015/f/fe/HUST_yl2.0.png"> | ||
| − | <div class="pizhu"> | + | <div class="pizhu">Figure 3: yeast two-hybrid principles</div> |
</div> | </div> | ||
| − | <p>To regulate DNA transcription by light, the system is based on a two-hybrid interaction in which a light-mediated protein interaction brings together two halves (a binding domain and an activation domain) of a split transcription factor of pGal. One of the light-mediated protein is Arabidopsis CIB1, a basic helix-loop-helix (bHLH) protein, and another is cryptochrome 2 (CRY2), a blue light-absorbing photosensor that binds CIB1 in its photoexcited state <sup>[3]</sup>. When applying the two-hybrid system to light control switch, there generated a couple of fusion protein with each half containing | + | <p>To regulate DNA transcription by light, the system is based on a two-hybrid interaction in which a light-mediated protein interaction brings together two halves (a binding domain and an activation domain) of a split transcription factor of pGal. One of the light-mediated protein is Arabidopsis CIB1, a basic helix-loop-helix (bHLH) protein, and another is cryptochrome 2 (CRY2), a blue light-absorbing photosensor that binds CIB1 in its photoexcited state <sup>[3]</sup>. When applying the two-hybrid system to light control switch, there generated a couple of fusion protein with each half containing a light-mediated part and transcription-factor-related part. If we remove the stimulation of blue light, dark reversion of CRY2 will dissociate its interaction with CIB1 and halt Gal4-dependent transcription. These modules require no exogenous chromophore, and are reversible within minutes thus triggering protein translocation at a sub-second time scale, and it even allow potential use in vivo in whole organisms (shown in Fig 3). </p> |
| − | <div > | + | <div> |
<img class="picture" src="https://static.igem.org/mediawiki/2015/2/26/Hust_figure4.png"> | <img class="picture" src="https://static.igem.org/mediawiki/2015/2/26/Hust_figure4.png"> | ||
| − | <div class="pizhu"> | + | <div class="pizhu">Figure 4: Light control part</div> |
</div> | </div> | ||
| − | <p>In the circuit, the two halves, CIB1 and CRY2 are fused separately with activation domain (AD) of Gal4 and DNA binding domain (BD) of Gal4,the fusion of which are respectively expressed by constitutive promoters pADH1 and pTEF. Both of them are essential to yeast genetics as the trigger of transcription requires their attachment to the promoter sequence at the same time. Commonly, it is almost impossible because they are too far from each other. But when exposed to blue light, such interaction of CIB1 and CRY2 brings together the split Gal1(similar to Gal4) transcription factor to control DNA transcription. As BD combines to the upstream activation sequence (UAS) and AD is successfully targeted to activate the promoter, downstream gene are able to be transcribed and translated, following the principle of yeast two-hybrid(shown in | + | <p>In the circuit, the two halves, CIB1 and CRY2 are fused separately with activation domain (AD) of Gal4 and DNA binding domain (BD) of Gal4,the fusion of which are respectively expressed by constitutive promoters pADH1 and pTEF. Both of them are essential to yeast genetics as the trigger of transcription requires their attachment to the promoter sequence at the same time. Commonly, it is almost impossible because they are too far from each other. But when exposed to blue light, such interaction of CIB1 and CRY2 brings together the split Gal1(similar to Gal4) transcription factor to control DNA transcription. As BD combines to the upstream activation sequence (UAS) and AD is successfully targeted to activate the promoter, downstream gene are able to be transcribed and translated, following the principle of yeast two-hybrid(shown in Fig 4).</p> |
<h3>Logic reverse control</h3><br> | <h3>Logic reverse control</h3><br> | ||
| − | <div > | + | <div> |
<img class="picture" src="https://static.igem.org/mediawiki/2015/3/3c/Hust_figure5.png"> | <img class="picture" src="https://static.igem.org/mediawiki/2015/3/3c/Hust_figure5.png"> | ||
| − | <div class="pizhu">Figure | + | <div class="pizhu">Figure 5:Logic reverse control</div> |
| − | + | ||
| − | + | ||
| − | + | ||
</div> | </div> | ||
| + | <p>Then a couple of promoter Panb1 and its inhibitory protein ROX1 <sup>[4]</sup> are set afterwards (shown in Figure 5). Actually, a light-activated promoter as we described above is still one step away from our needs. Since the applications of this project are expected to be put into marine environment, it is highly possible that the marine yeasts are unable to get to sunlight in most of the blue water region. Based on the above, we generated a logic reverse control. After introducing Panb1 and ROX1, the system’s function is exquisitely switched: in the condition of light, ROX1 in the downstream of pGal1 are expressed to inhibit expressing our functional proteins. Only total darkness can remove the inhibition and allow the expression of target protein in abysmal sea of all time.</p> | ||
| + | </div> | ||
| − | + | <div class="description"><a name="supporting"></a><br><br> | |
<div class="dongxi"></div> | <div class="dongxi"></div> | ||
| − | <h2 style="color:black" align="left"><b>3.Supporting | + | <h2 style="color:black" align="left"><b>3.Supporting System</b></h2> |
| − | <p>Next, to preserve the kit’s function, we placed a silica binding protein Si-tag right after Panb1. It’s not for direct consolidation, but to target marine yeasts to the surface of sands. Considering waves may occur occasionally underwater, a guarantee for the yeasts not to flow away is really necessary, which equals to immobilize them on silicon. When we fused an anchor protein YLcwp3 at the C-terminal of Si-tag and a signal peptide LIP2 prepro at the N-terminal | + | <p>Next, to preserve the kit’s function, we placed a silica binding protein Si-tag right after Panb1. It’s not for direct consolidation, but to target marine yeasts to the surface of sands. Considering waves may occur occasionally underwater, a guarantee for the yeasts not to flow away is really necessary, which equals to immobilize them on silicon. When we fused an anchor protein YLcwp3 at the C-terminal of Si-tag and a signal peptide LIP2 prepro at the N-terminal of Si-tag, we can display Si-tag onto the surface of cell wall and link the yeasts’ fate to sands thoroughly.</p> |
| − | <div > | + | <div> |
| − | <img class="picture" src="https://static.igem.org/mediawiki/2015/8/8d/Hust_figure6.png"> | + | <img style="width:500px;height:300px;float:left" class="picture" src="https://static.igem.org/mediawiki/2015/8/8d/Hust_figure6.png"> |
| − | + | <img style="width:300px;height:300px;float:left" class="picture" src="https://static.igem.org/mediawiki/2015/1/15/Hust_2.png"> | |
| − | <div class="pizhu">Figure | + | <div class="pizhu">Figure 6: Supporting system:displaying Si-tag on the cell wall of Y.L</div> |
</div> | </div> | ||
| − | <h3>Si-tag</h3><br> | + | <h3 style="color:black;margin-top:30px" align="left">Si-tag</h3><br> |
| − | <p>Si-tag is 50S ribosomal protein L2 in the genome of E.coli<sup>[5,6]</sup> . It has been found from several bacterial strains and proved to have a silica-binding property. It is demonstrated that an L2-protein fusion binds to silica surfaces 20- to 100-fold more strongly than poly-Arg-tagged proteins, showing its strong superiority in silica binding. The Si-tag consists of three domains. | + | <p style="width:500px;float:left" >Si-tag is 50S ribosomal protein L2 in the genome of E.coli<sup>[5,6]</sup> . It has been found from several bacterial strains and proved to have a silica-binding property. It is demonstrated that an L2-protein fusion binds to silica surfaces 20- to 100-fold more strongly than poly-Arg-tagged proteins, showing its strong superiority in silica binding. The Si-tag consists of three domains. Based on previous study, it is found that domain 1 and 3 of recombinant Si-tag bind much stronger than domain 2. Following this result, we performed further study to see how the combinations of different domains work in our surface display system. Indeed, we find it meaningful as a kit that fits in several circumstances. This varying intensity of viscosity is just what we want as a kit to be to utilized following different requirement. In our study, the separate ones, domain 1, 2, 3, and combinations as fusion domains1+2, 1+3, 2+3, 1+2+3, are all tested. Hoping a linker could bring better flexibility between two domains, we also put a short GS linker between domain 1&3, creating the 1+Linker+3 Si-tag.</p> |
| − | + | <img style="width:320px;float:left;height:400px;margin-left:20px;" src="https://static.igem.org/mediawiki/2015/6/6c/Description-Si-func.png"/> | |
| − | <p>Ylcwp3 is the covalently bound GPI-anchored cell wall protein of Yarrowia lipolytica. The protein allows the target to be covalently bound to β-1, 6-glucan of the cell wall. The nucleotide sequence encoding 110 amino acids of the Ylcwp3 C terminus has been widely utilized for the construction of the cell surface display vector in Y. Lipolytica. In previous study, the observed high activity of the immobilized lipase LIP2 on the surface clearly shows the efficiency of the surface display system<sup>[7,9]</sup>.Therefore we adopt YLcwp3 as an anchor to target the interest protein on the surface of the cell wall.</p> | + | |
| − | + | <div style="color:black;margin-top:30px" align="left"><b>Anchor Protein</b></div><br> | |
| − | <p>Yarrowia.lipolytica naturally secretes lipases LIP2. The signal peptide LIP2 prepro we employed is a modified signal peptide from Y.lipolytica LIP2 with the sequence consisting 13 aa pre, 4 XA or XP dipeptides, 10 aa pro and KR cleavage site. It can leads the flocculating protein Mcfp-3 to be secreted out of cell to perform its flocculating function, during the secretion progress, the signal peptide will be deleted in the Golgi apparatus to eliminate its interference to the secreting protein<sup>[8,9]</sup> . | + | <p style="width:820px;float:left" >Ylcwp3 is the covalently bound GPI-anchored cell wall protein of Yarrowia lipolytica. The protein allows the target to be covalently bound to β-1, 6-glucan of the cell wall. The nucleotide sequence encoding 110 amino acids of the Ylcwp3 C terminus has been widely utilized for the construction of the cell surface display vector in Y. Lipolytica. In previous study, the observed high activity of the immobilized lipase LIP2 on the surface clearly shows the efficiency of the surface display system<sup>[7,9]</sup>.Therefore we adopt YLcwp3 as an anchor to target the interest protein on the surface of the cell wall.<br><br><br></p> |
| − | + | <div style="color:black;margin-top:30px" align="left"><b>Signal Peptide</b></div><br> | |
| − | + | <p style="width:500px;float:left" >Yarrowia.lipolytica naturally secretes lipases LIP2. The signal peptide LIP2 prepro we employed is a modified signal peptide from Y.lipolytica LIP2 with the sequence consisting 13 aa pre, 4 XA or XP dipeptides, 10 aa pro and KR cleavage site. It can leads the flocculating protein Mcfp-3 to be secreted out of cell to perform its flocculating function, during the secretion progress, the signal peptide will be deleted in the Golgi apparatus to eliminate its interference to the secreting protein<sup>[8,9]</sup> . | |
| − | + | </p><img style="width:320px;float:left;height:250px;margin-left:20px;" src="https://static.igem.org/mediawiki/2015/d/d1/Description-surf-diplay.png"/> | |
| − | + | </div> | |
| + | |||
| + | <div class="description"><a name="flocculating"></a><br><br> | ||
<div class="dongxi"></div> | <div class="dongxi"></div> | ||
| − | <h2 style="color:black" align="left"><b>4.Flocculating | + | <h2 style="color:black" align="left"><b>4.Flocculating System</b></h2> |
| − | <div > | + | <div> |
| − | <img class="picture" src="https://static.igem.org/mediawiki/2015/8/89/Hust_figure7.png"> | + | <img style="width:500px;height:300px;float:left" class="picture" src="https://static.igem.org/mediawiki/2015/8/89/Hust_figure7.png"> |
| − | + | <img style="width:300px;height:300px;float:left" class="picture" src="https://static.igem.org/mediawiki/2015/f/f5/Hust_3.png"> | |
| − | <div class="pizhu">Figure 7 Flocculating system:Secreting Mcfp-3 among sand | + | <div class="pizhu">Figure 7: Flocculating system:Secreting Mcfp-3 among sand</div> |
| − | </div> | + | </div> |
| − | + | <p>Following is flocculating agent secretion circuit which briefly consists of the promoter anb1, LIP2 prepro signal peptide, Mcfp-3 flocculating protein and its terminator yADH1. Mcfp-3 is the main flocculating material. It can perform self-assembly and thus get high affinity to silica and other solid surface. So the secreted Mcfp-3 protein can cross link sand particles together and complete flocculation.</p> | |
| − | <p> | + | |
<h3>Flocculating agent</h3><br> | <h3>Flocculating agent</h3><br> | ||
| − | <p>Mcfp-3 is the filopodia protein secreted from Mytilus californianus<sup>[10]</sup> . The protein is important to the formation of filopodia to help mussels permanently or temporarily tether to the rough surface of reef or ship-body. This coherent substance shows an excellent adhesion performance with no substitute under water for sustaining in the repeating wash of waves and having no any | + | <p style="width:500px;float:left" >Mcfp-3 is the filopodia protein secreted from Mytilus californianus<sup>[10]</sup> . The protein is important to the formation of filopodia to help mussels permanently or temporarily tether to the rough surface of reef or ship-body. This coherent substance shows an excellent adhesion performance with no substitute under water for sustaining in the repeating wash of waves and having no any known toxicity. The properties are owing to the amino acid content which are successively DOPA (28mol%), glycine, lysine and asparagine. The presence of DOPA in the foot protein suggests a role in protein-protein and protein-substrate(glass eg.) interactions related to the formation of adhesive plaques. The electron rich nature of DOPA and its susceptibility to oxidation can lead to the formation of o-quinone with subsequent tanning or sclerotization to form a cross-linked stable products. Furthermore, it is noteworthy that the protein shows an intrinsic ability of self-assemble into stable, higher molecular weight forms that may further enhance our consolidation function<sup>[11]</sup>.</p> |
| − | + | <img style="width:300px;float:left;height:250px;margin-left:40px;" src="https://static.igem.org/mediawiki/2015/d/d8/Description-floccu-combi.png"/> | |
| − | + | </div> | |
| − | + | <div class="description"><a name="host"></a><br><br> | |
| − | + | <div class="dongxi"></div> | |
| − | + | <h2 style="color:black" align="left"><b>5. Yarrowia Lipolytca</b></h2> | |
| − | + | <p style="width:500px;float:left" >We adopt a marine yeast Yarrowia lipolytca JMY1212 as our chassis <sup>[12]</sup> for the specific application this year: building artificial reef. Our host is generally regarded as safe microorganism and has an abroad application prospect in industries of foods and medicine. Researches show that Y.lipolytica is widely found in marine environment and exists stably in the ocean and healthy marine fishes’ intestine. It is able to make use of Glucose, ethanol, acetate or other hydrophobic substrate (alkanes, fatty acid and lipid ) as carbon source with fast growing capacity and adaptability for high-density fermentation. Moreover in the expression system of Y.lipolytica, transformed plasmids can integrate into multiple site and they are all genetically stable in the genome. And the signal peptide XPR2 pre, LIP2 prepro and so on are all demonstrated to own high efficiency of secreting protein. So it is obvious Y.lipolytica is a powerful host to express and secrete complicated recombinant protein. Based on the above mentioned reasons, Yarrowia lipolytica are regarded as an ideal strain for our marine application of micro-consolidation.</p> | |
| − | < | + | <img style="width:300px;float:left;margin-left:20px;" src="https://static.igem.org/mediawiki/2015/e/ea/Hust_4.png"/> |
| − | <img | + | |
</div> | </div> | ||
| − | + | <div class="description"><a name="improvement"></a><br><br> | |
| − | + | ||
| − | + | ||
| − | + | ||
<div class="dongxi"></div> | <div class="dongxi"></div> | ||
| − | + | <h2 style="color:black" align="left"><b>6.Biobrick Improvement</b></h2> | |
| − | < | + | |
| − | + | <p style="width:500px;float:left">In the early period of our project design, we found two SpeI illegal sites in a Part BBa_K191007 when glancing over parts Registered in pools. Though we wouldn't use this part in our project, we still removed its illegal sites by site-directed mutagenesis, in order to make it meet the requirements of RFC10 and more convenient to be assembled. According to the reference <sup>[12]</sup>,we successfully found the appropriate sites for mutation without affecting the part’s function.<br> | |
| + | Finally, we created 3 different mutants of Part <a href="http://parts.igem.org/Part:BBa_K191007">BBa_K191007</a>. In order to test and verify our mutants’ function, we inserted RBS <a href="http://parts.igem.org/Part:BBa_B0032">(BBa_B0032)</a> + GFP <a href="http://parts.igem.org/Part:BBa_E0040">(BBa_E0040)</a> into the downstream of them. We have intended to measure their fluorescence value. However,unfortunately, we had some trouble in getting the tryptophan analog, 3-indoleacrylicacid, a competitive inhibitor of tryptophan in the formation of the specific aporepressor, so we couldn't complete the verification work. We hope that their verification can be finished in the future.<br> | ||
| + | Moreover, we also improved the characterization of five existing BioBrick Parts. They are <a href="http://parts.igem.org/Part:BBa_K207001">BBa_K207001</a> | ||
| + | , | ||
| + | <a href="http://parts.igem.org/Part:BBa_K1144001">BBa_K1144001</a> | ||
| + | , | ||
| + | <a href="http://parts.igem.org/Part:BBa_K105007">BBa_K105007</a> | ||
| + | , | ||
| + | <a href="http://parts.igem.org/Part:BBa_K950002">BBa_K950002</a> | ||
| + | , | ||
| + | and | ||
| + | <a href="http://parts.igem.org/Part:BBa_K950001">BBa_K950001</a>.</p> | ||
| + | <img style="width:300px;float:left;margin-left:20px;" class="picture" src="https://static.igem.org/mediawiki/2015/0/07/Hust_5.png"> | ||
</div> | </div> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | <table border="1"> | + | <div class="description"> |
| + | <table border="1" style="width:810px;height:auto"> | ||
<tr> | <tr> | ||
<th>Type</th> | <th>Type</th> | ||
| Line 484: | Line 491: | ||
<!--reference--> | <!--reference--> | ||
| − | <div class="description"><a name=" | + | <div class="description"><a name="reference"></a><br><br> |
<div class="dongxi"></div> | <div class="dongxi"></div> | ||
| − | <h2 style="color:black" align="left"><b> | + | <h2 style="color:black" align="left"><b>Reference</b></h2> |
<p> | <p> | ||
[1] Braselmann S, Graninger P, Busslinger M. A selective transcriptional induction system for mammalian cells based on Gal4-estrogen receptor fusion proteins[J]. Proceedings of the National Academy of Sciences, 1993, 90(5): 1657-1661.<br> | [1] Braselmann S, Graninger P, Busslinger M. A selective transcriptional induction system for mammalian cells based on Gal4-estrogen receptor fusion proteins[J]. Proceedings of the National Academy of Sciences, 1993, 90(5): 1657-1661.<br> | ||
| Line 501: | Line 508: | ||
[12]Madzak C, Gaillardin C, Beckerich J M. Heterologous protein expression and secretion in the non-conventional yeast Yarrowia lipolytica: a review[J]. Journal of Biotechnology, 2004, 109(1): 63-81.<br> | [12]Madzak C, Gaillardin C, Beckerich J M. Heterologous protein expression and secretion in the non-conventional yeast Yarrowia lipolytica: a review[J]. Journal of Biotechnology, 2004, 109(1): 63-81.<br> | ||
[13] Yanofsky C, Platt T, Crawford I P, et al. The complete nucleotide sequence of the tryptophan operon of Escherichia coli [J]. Nucleic acids research, 1981, 9(24): 6647-6668. | [13] Yanofsky C, Platt T, Crawford I P, et al. The complete nucleotide sequence of the tryptophan operon of Escherichia coli [J]. Nucleic acids research, 1981, 9(24): 6647-6668. | ||
| − | </p> | + | <br><br><br></p> |
</div> | </div> | ||
Latest revision as of 16:58, 18 September 2015

click it~


According to requirement of iGEM wiki construction, we hosted our cartoon instruction on iGEM server. For those can not play this video on your Browser such as IE11, please turn to view our wiki with google chrome or directly donwload this video HERE from iGEM server.
1.Description
In our project of Euk.cement, we employ Yarrowia lipolytica JMY1212 as our chassis and generate three circuits respectively. We use expression vector p414TEF to load the light control part of darkness induction system and JMP62 as backbone to carry the negative control part together with flocculating part and supporting part. The whole project is aiming to consolidate sands or soil particles more flexibly in proper way, time and region. Next, we will show all the details.

As mentioned above, our design is briefly divided into three parts: Darkness induction system, flocculating system and supporting system. The latter two are both under control of the former one with the same upstream promoter Anb1. Fusion protein CRY2-BD and CIB1-AD are respectively expressed by constitutive promoter pTEF and pADH1. CRY2-BD has high affinity to bind CIB1-AD only when induced by blue light (even contained in nature sunlight) . This interaction can activate pGal1 owing to the combination between CIB1 and CRY2’s special structures thus induce the downstream promoter pGal1 and express ROX1 gene. Then expressed ROX1 protein can inhibit the promoter Anb1. So our engineered circuit is turned off when exposed to light. On the contrary, in darkness, the structural-reversed CRY2 will dissociate with CIB1. Without CRY2-BD’s target function, AD is hard to combine and active pGal1 in the complex environment of nucleus, so the downstream ROX1 is no longer transcribed. With the degradation of translated ROX1 protein, pAnb1 starts to express the downstream gene as the supporting matter and flocculating agent :LIP2 pro-Si-tag-YLcwp3 and LIP2-pro-Mcfp-3 fusion proteins(shown in Fig 1).
2.Darkness Induction System


The first one is the expression control system. Each system should apply a kind of trigger of working system. While a number of inducible systems are available for controlling protein expression in yeast, almost none of them make sense for us, as our project’s practical applications are currently considered to exist mainly in marine environment, which makes the amount of chemical inducers to be used a huge burden.
So we give up several common promoters including those naturally responsive to molecules such as galactose, methionine, and copper, or engineered regulatory systems that respond to orthogonal molecules such as estrogen (Gal-ER-VP16)[1] or doxycycline (Tet-OFF)[2] .In fact, some of them are really robust and well-designed pathways. However, we find a better method instead, a darkness induction system based on light control yeast two-hybrid principles, because darkness is the most common environment factor that our Euk.cement cells will face in working.
Our original idea comes from dimers that allow inducible control of protein-protein interactions, which are powerful tools for manipulating biological processes. On this basis, we successfully utilized a split transcription factor that is reconstituted by light-dependent protein–protein interactions as in yeast two-hybrid systems.
Compared to those chemical-genetic tools, the light control system not only meets our demand, but also possesses many other advantages. Despite the general success of chemical inducers, the potential problems, such as toxic, nonspecific, or pleiotropic effects of these treatments can impose limitations on their use in sensitive chassis or environment. Also, light control is far more fast, precise, low cost and easy. It even allows exquisite dose-dependent control over protein levels, as light can be delivered for defined amounts of time and be instantly removed.
Principle of light control yeast two hybrid

To regulate DNA transcription by light, the system is based on a two-hybrid interaction in which a light-mediated protein interaction brings together two halves (a binding domain and an activation domain) of a split transcription factor of pGal. One of the light-mediated protein is Arabidopsis CIB1, a basic helix-loop-helix (bHLH) protein, and another is cryptochrome 2 (CRY2), a blue light-absorbing photosensor that binds CIB1 in its photoexcited state [3]. When applying the two-hybrid system to light control switch, there generated a couple of fusion protein with each half containing a light-mediated part and transcription-factor-related part. If we remove the stimulation of blue light, dark reversion of CRY2 will dissociate its interaction with CIB1 and halt Gal4-dependent transcription. These modules require no exogenous chromophore, and are reversible within minutes thus triggering protein translocation at a sub-second time scale, and it even allow potential use in vivo in whole organisms (shown in Fig 3).

In the circuit, the two halves, CIB1 and CRY2 are fused separately with activation domain (AD) of Gal4 and DNA binding domain (BD) of Gal4,the fusion of which are respectively expressed by constitutive promoters pADH1 and pTEF. Both of them are essential to yeast genetics as the trigger of transcription requires their attachment to the promoter sequence at the same time. Commonly, it is almost impossible because they are too far from each other. But when exposed to blue light, such interaction of CIB1 and CRY2 brings together the split Gal1(similar to Gal4) transcription factor to control DNA transcription. As BD combines to the upstream activation sequence (UAS) and AD is successfully targeted to activate the promoter, downstream gene are able to be transcribed and translated, following the principle of yeast two-hybrid(shown in Fig 4).
Logic reverse control

Then a couple of promoter Panb1 and its inhibitory protein ROX1 [4] are set afterwards (shown in Figure 5). Actually, a light-activated promoter as we described above is still one step away from our needs. Since the applications of this project are expected to be put into marine environment, it is highly possible that the marine yeasts are unable to get to sunlight in most of the blue water region. Based on the above, we generated a logic reverse control. After introducing Panb1 and ROX1, the system’s function is exquisitely switched: in the condition of light, ROX1 in the downstream of pGal1 are expressed to inhibit expressing our functional proteins. Only total darkness can remove the inhibition and allow the expression of target protein in abysmal sea of all time.
3.Supporting System
Next, to preserve the kit’s function, we placed a silica binding protein Si-tag right after Panb1. It’s not for direct consolidation, but to target marine yeasts to the surface of sands. Considering waves may occur occasionally underwater, a guarantee for the yeasts not to flow away is really necessary, which equals to immobilize them on silicon. When we fused an anchor protein YLcwp3 at the C-terminal of Si-tag and a signal peptide LIP2 prepro at the N-terminal of Si-tag, we can display Si-tag onto the surface of cell wall and link the yeasts’ fate to sands thoroughly.


Si-tag
Si-tag is 50S ribosomal protein L2 in the genome of E.coli[5,6] . It has been found from several bacterial strains and proved to have a silica-binding property. It is demonstrated that an L2-protein fusion binds to silica surfaces 20- to 100-fold more strongly than poly-Arg-tagged proteins, showing its strong superiority in silica binding. The Si-tag consists of three domains. Based on previous study, it is found that domain 1 and 3 of recombinant Si-tag bind much stronger than domain 2. Following this result, we performed further study to see how the combinations of different domains work in our surface display system. Indeed, we find it meaningful as a kit that fits in several circumstances. This varying intensity of viscosity is just what we want as a kit to be to utilized following different requirement. In our study, the separate ones, domain 1, 2, 3, and combinations as fusion domains1+2, 1+3, 2+3, 1+2+3, are all tested. Hoping a linker could bring better flexibility between two domains, we also put a short GS linker between domain 1&3, creating the 1+Linker+3 Si-tag.

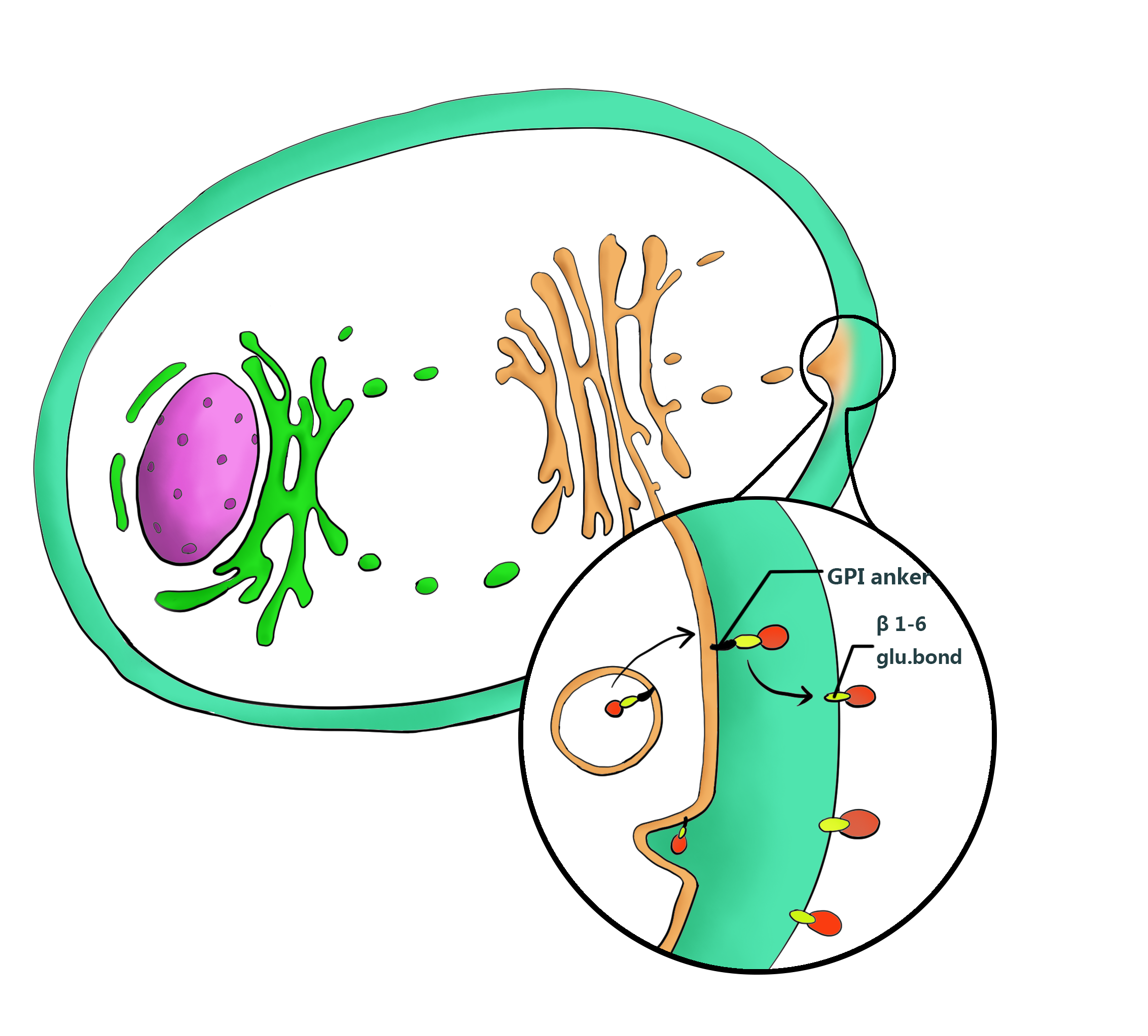
Ylcwp3 is the covalently bound GPI-anchored cell wall protein of Yarrowia lipolytica. The protein allows the target to be covalently bound to β-1, 6-glucan of the cell wall. The nucleotide sequence encoding 110 amino acids of the Ylcwp3 C terminus has been widely utilized for the construction of the cell surface display vector in Y. Lipolytica. In previous study, the observed high activity of the immobilized lipase LIP2 on the surface clearly shows the efficiency of the surface display system[7,9].Therefore we adopt YLcwp3 as an anchor to target the interest protein on the surface of the cell wall.
Yarrowia.lipolytica naturally secretes lipases LIP2. The signal peptide LIP2 prepro we employed is a modified signal peptide from Y.lipolytica LIP2 with the sequence consisting 13 aa pre, 4 XA or XP dipeptides, 10 aa pro and KR cleavage site. It can leads the flocculating protein Mcfp-3 to be secreted out of cell to perform its flocculating function, during the secretion progress, the signal peptide will be deleted in the Golgi apparatus to eliminate its interference to the secreting protein[8,9] .
4.Flocculating System


Following is flocculating agent secretion circuit which briefly consists of the promoter anb1, LIP2 prepro signal peptide, Mcfp-3 flocculating protein and its terminator yADH1. Mcfp-3 is the main flocculating material. It can perform self-assembly and thus get high affinity to silica and other solid surface. So the secreted Mcfp-3 protein can cross link sand particles together and complete flocculation.
Flocculating agent
Mcfp-3 is the filopodia protein secreted from Mytilus californianus[10] . The protein is important to the formation of filopodia to help mussels permanently or temporarily tether to the rough surface of reef or ship-body. This coherent substance shows an excellent adhesion performance with no substitute under water for sustaining in the repeating wash of waves and having no any known toxicity. The properties are owing to the amino acid content which are successively DOPA (28mol%), glycine, lysine and asparagine. The presence of DOPA in the foot protein suggests a role in protein-protein and protein-substrate(glass eg.) interactions related to the formation of adhesive plaques. The electron rich nature of DOPA and its susceptibility to oxidation can lead to the formation of o-quinone with subsequent tanning or sclerotization to form a cross-linked stable products. Furthermore, it is noteworthy that the protein shows an intrinsic ability of self-assemble into stable, higher molecular weight forms that may further enhance our consolidation function[11].

5. Yarrowia Lipolytca
We adopt a marine yeast Yarrowia lipolytca JMY1212 as our chassis [12] for the specific application this year: building artificial reef. Our host is generally regarded as safe microorganism and has an abroad application prospect in industries of foods and medicine. Researches show that Y.lipolytica is widely found in marine environment and exists stably in the ocean and healthy marine fishes’ intestine. It is able to make use of Glucose, ethanol, acetate or other hydrophobic substrate (alkanes, fatty acid and lipid ) as carbon source with fast growing capacity and adaptability for high-density fermentation. Moreover in the expression system of Y.lipolytica, transformed plasmids can integrate into multiple site and they are all genetically stable in the genome. And the signal peptide XPR2 pre, LIP2 prepro and so on are all demonstrated to own high efficiency of secreting protein. So it is obvious Y.lipolytica is a powerful host to express and secrete complicated recombinant protein. Based on the above mentioned reasons, Yarrowia lipolytica are regarded as an ideal strain for our marine application of micro-consolidation.

6.Biobrick Improvement
In the early period of our project design, we found two SpeI illegal sites in a Part BBa_K191007 when glancing over parts Registered in pools. Though we wouldn't use this part in our project, we still removed its illegal sites by site-directed mutagenesis, in order to make it meet the requirements of RFC10 and more convenient to be assembled. According to the reference [12],we successfully found the appropriate sites for mutation without affecting the part’s function.
Finally, we created 3 different mutants of Part BBa_K191007. In order to test and verify our mutants’ function, we inserted RBS (BBa_B0032) + GFP (BBa_E0040) into the downstream of them. We have intended to measure their fluorescence value. However,unfortunately, we had some trouble in getting the tryptophan analog, 3-indoleacrylicacid, a competitive inhibitor of tryptophan in the formation of the specific aporepressor, so we couldn't complete the verification work. We hope that their verification can be finished in the future.
Moreover, we also improved the characterization of five existing BioBrick Parts. They are BBa_K207001
,
BBa_K1144001
,
BBa_K105007
,
BBa_K950002
,
and
BBa_K950001.

| Type | Part Number | Description | Length(bp) |
|---|---|---|---|
| Regulatory | BBa_K1592020 | Part mutant1 | 85 |
| Regulatory | BBa_K1592021 | Part mutant2 | 85 |
| Regulatory | BBa_K1592022 | Part mutant3 | 85 |
| Composite | BBa_K1592023 | Part mutant1+RBS+GFP | 832 |
| Composite | BBa_K1592024 | Part mutant2+RBS+GFP | 832 |
| Composite | BBa_K1592025 | Part mutant3+RBS+GFP | 832 |
Reference
[1] Braselmann S, Graninger P, Busslinger M. A selective transcriptional induction system for mammalian cells based on Gal4-estrogen receptor fusion proteins[J]. Proceedings of the National Academy of Sciences, 1993, 90(5): 1657-1661.
[2]Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies[J]. Science, 2005, 310(5747): 486-489.
[3]Hughes R M, Bolger S, Tapadia H, et al. Light-mediated control of DNA transcription in yeast[J]. Methods, 2012, 58(4): 385-391.
[4] Balasubramanian B, Lowry C V, Zitomer R S. The Rox1 repressor of the Saccharomyces cerevisiae hypoxic genes is a specific DNA-binding protein with a high-mobility-group motif[J]. Molecular and cellular biology, 1993, 13(10): 6071-6078.
[5]Taniguchi K, Nomura K, Hata Y, et al. The Si‐tag for immobilizing proteins on a silica surface[J]. Biotechnology and bioengineering, 2007, 96(6): 1023-1029.
[6]Ikeda T, Hata Y, Ninomiya K, et al. Oriented immobilization of antibodies on a silicon wafer using Si-tagged protein A[J].Analytical biochemistry, 2009, 385(1): 132-137.
[7]Yuzbasheva E Y, Yuzbashev T V, Laptev I A, et al. Efficient cell surface display of Lip2 lipase using C-domains of glycosylphosphatidylinositol-anchored cell wall proteins of Yarrowia lipolytica[J]. Applied microbiology and biotechnology, 2011, 91(3): 645-654.
[8]Nicaud J M, Madzak C, Broek P, et al. Protein expression and secretion in the yeast Yarrowia lipolytica[J]. FEMS yeast research, 2002, 2(3): 371-379.
[9]Madzak C, Gaillardin C, Beckerich J M. Heterologous protein expression and secretion in the non-conventional yeast Yarrowia lipolytica: a review[J]. Journal of Biotechnology, 2004, 109(1): 63-81.
[10]Platko J D, Deeg M, Thompson V, et al. Heterologous expression of Mytilus californianus foot protein three (Mcfp-3) in Kluyveromyces lactis[J]. Protein expression and purification, 2008, 57(1): 57-62.
[11]Zhong C, Gurry T, Cheng A A, et al. Strong underwater adhesives made by self-assembling multi-protein nanofibres[J]. Nature nanotechnology, 2014.
[12]Madzak C, Gaillardin C, Beckerich J M. Heterologous protein expression and secretion in the non-conventional yeast Yarrowia lipolytica: a review[J]. Journal of Biotechnology, 2004, 109(1): 63-81.
[13] Yanofsky C, Platt T, Crawford I P, et al. The complete nucleotide sequence of the tryptophan operon of Escherichia coli [J]. Nucleic acids research, 1981, 9(24): 6647-6668.