|
|
| (45 intermediate revisions by 3 users not shown) |
| Line 2: |
Line 2: |
| | <html> | | <html> |
| | <!-- 侧边导航 --> | | <!-- 侧边导航 --> |
| | + | <div class="sui-highlight" style="width: 100%; height: 2px; background-color: #D7E79F; margin-bottom: 50px;"></div> |
| | <div class="sui-side-nav"> | | <div class="sui-side-nav"> |
| | <ul> | | <ul> |
| | + | <li><a href="#Abstract">Abstract</a></li> |
| | <li><a href="#Background">Background</a> | | <li><a href="#Background">Background</a> |
| | <ul class="sub-ul"><!-- 第二级菜单 --> | | <ul class="sub-ul"><!-- 第二级菜单 --> |
| | <li><a href="#Meanings-of-Timer-in-Microbes">Meanings of Timer in Microbes</a></li> | | <li><a href="#Meanings-of-Timer-in-Microbes">Meanings of Timer in Microbes</a></li> |
| − | <li><a href="#Ideas-of-Timer">Ideas of Timer</a></li>
| |
| | <li><a href="#Recombinase-System">Recombinase System</a></li> | | <li><a href="#Recombinase-System">Recombinase System</a></li> |
| | </ul> | | </ul> |
| | </li> | | </li> |
| − | <li><a href="#Abstract">Abstract</a></li> | + | |
| − | <li><a href="#Overview">Overview</a> | + | <li><a href="#Description">Description</a> |
| − | <ul class="sub-ul">
| + | <ul class="sub-ul"><!-- 第二级菜单 --> |
| − | <li><a href="#Matching-and-Testing">Matching and Testing</a></li>
| + | <li><a href="#Matching-and-Testing">Matching and Testing</a></li> |
| − | <li><a href="#Bacteria-Timer">Bacteria Timer</a></li>
| + | <li><a href="#Prokaryotic-Timer">Prokaryotic Timer</a></li> |
| − | <li><a href="#Yeast-Timer">Yeast Timer</a></li>
| + | <li><a href="#Telomeric-Timer">Telomeric Timer</a></li> |
| − | <li><a href="#Modelling">Modelling</a><li></li>
| + | <li><a href="#Eukaryotic-Timer">Eukaryotic Timer</a></li> |
| − | </ul>
| + | <li><a href="#Registry-Contribution">Registry Contribution</a></li> |
| − | </li>
| + | |
| − | <li> <a href="#Collaboration">Collaboration</a>
| + | |
| − | <ul class="sub-ul">
| + | |
| − | <li><a href="#Collaboration-with-ZJU">Collaboration with ZJU</a></li> | + | |
| − | <li><a href="#Collaboration-with-SJTU">Collaboration with SJTU</a></li> | + | |
| − | <li><a href="#Collaboration-with-THU">Collaboration with ZTH</a></li>
| + | |
| | </ul> | | </ul> |
| − | </li>
| + | </li> |
| | </ul> | | </ul> |
| | </div> | | </div> |
| Line 34: |
Line 29: |
| | | | |
| | <div class="sui-article"> | | <div class="sui-article"> |
| − | | + | <div id="Abstract" class="scrollto"> |
| | + | <h1>Abstract</h1> |
| | + | <p>When we talk about internal clock, we actually refer to the circadian rhythm of higher animals and plants. Sensing the cycling environmental factors (light and temperature, etc.), circadian clock allows organism to behave accordingly and reguarly. This year, we develop a plug-in for microbes, a system that can build up an internal clock for microbes and guide them to live regularly as well as to work efficiently. Also, we could insert this system into dangerous strains and endue life span, they are doomed to die before causing any biohazard. Flip units based on recombinase system are introduced in our system. One flip unit is able to calculate a specific period of time, while several flip units form one time cycle. So, when we combine several time cycles together, a timer is constructed. Different flip unit assemblies creat distinctive time cycles, forms different kinds of timers. We believe this system will be of great significance to biological devices with time-related concepts.</p> |
| | + | </div> |
| | <div id="Background" class="scrollto"> | | <div id="Background" class="scrollto"> |
| | <h1>Background</h1> | | <h1>Background</h1> |
| − |
| |
| | <div id="Meanings-of-Timer-in-Microbes" class="scrollto"> | | <div id="Meanings-of-Timer-in-Microbes" class="scrollto"> |
| | <h2>Meanings of Timer in Microbes</h2> | | <h2>Meanings of Timer in Microbes</h2> |
| − | <p>It is well known that the internal clock is of great significance to human life. The time rhythm let people work and rest regularly, which keeps us healthy and energetic. However, in microbes, especially procaryotic organism, we could hardly find a strain with rhythm.</p> | + | <p>It is well known that the internal clock is of great significance to human life. The time rhythm let people work and rest regularly, which keeps us healthy and energetic. However, in microbes, especially prokaryotic organism, we could hardly find a strain with rhythm.</p> |
| − | <p>Actually, adding rhythm into microbes is a meaningful thing, by which would provide us a brand new technology platform or control system. This year, we hope to construct a rhythm device with time limit. As follows, we will use 3 simple hypothetical cases to illustrate its future application potentials.</p> | + | <p>Actually, adding rhythm into microbes is a meaningful thing, by which would provide us a brand new technology platform or control system. This year, we hope to construct a rhythm device with time limit. As follows, we will use 3 simple hypothetical cases to illustrate its future application potentials. |
| − | <p>a.Industry application of microbial rhythm<p>
| + | |
| − | <p>When it comes to microbial industry, we often find restrictive reaction space and strict operations necessary, since in the current industrial production, without autonomic consciousness, microorganisms could only behave under stress. So, if we want to express several proteins in turn, this limit determines that we have to add a new inducer once in a while to initiate the expression of a new gene. However, once microbes have autonomic rhythm and the concept of time, any operation with timing demands could be done in a autonomic way. We could make bacteria express proteins in chronological order, or realize certain behaviors by periodic expression, and then, apply to industrial mass production.</p>
| + | |
| − | <p>b.The advantages of microorganism rhythm in therapy </p>
| + | |
| − | <p>Many diseases are associated with human body's endocrine, and human endocrine usually has its own cycle. For a patient with insomnia, sleeping pills almost become daily necessities, which is inconvenient. However, using the engineered E.coli to secret sleeping pills analogues with rhythm, then these drugs would go through intestines into the systemic circulation to effect the neural system, may regulate the body's own endocrine in a longer period of time, in order to ease insomnia. In the same way, long-termed treatments become available for any diseases or symptoms associated with time.</p>
| + | |
| − | <p>c.The meaning of hereditary time limits (delayed suicide)</p>
| + | |
| − | <p>By far, many iGEM projects are based on the construction of strains for medical treatments, but planting living microorganisms into human body has great potential safety hazards. However, if the microbes have time limit devices, we would be able to use them, which were designed to commit suicide in calculated days, to treat human. Such microorganisms would die naturally after function, and their offspring will die too, which ensure the safety of human body. Time limits are also capable of more sophisticated applications, for example, timed initiation of gene expression. In the future biological industries and biomedical fields, the prospects for it are almost limitless.
| + | |
| | </p> | | </p> |
| | + | |
| | + | <br> |
| | + | <p>a. Industry application of microbial rhythm</p> |
| | + | <p>When it comes to microbial industry, we often find restrictive reaction space and strict operations necessary, since in the current industrial production, without autonomic consciousness, microorganisms could only behave under stress. So, if we want to express several proteins in turn, this limit determines that we have to add a new inducer once in a while to initiate the expression of a new gene. However, once microbes have autonomic rhythm and the concept of time, any operation with timing demands could be done in a autonomic way. We could make bacteria express proteins in chronological order, or realize certain behaviors by periodic expression, and then, apply to industrial mass production. |
| | + | </p> |
| | + | <img src="https://static.igem.org/mediawiki/2015/thumb/8/88/LdwBG1.jpeg/800px-LdwBG1.jpeg" alt=""> |
| | + | |
| | + | <br> |
| | + | <br> |
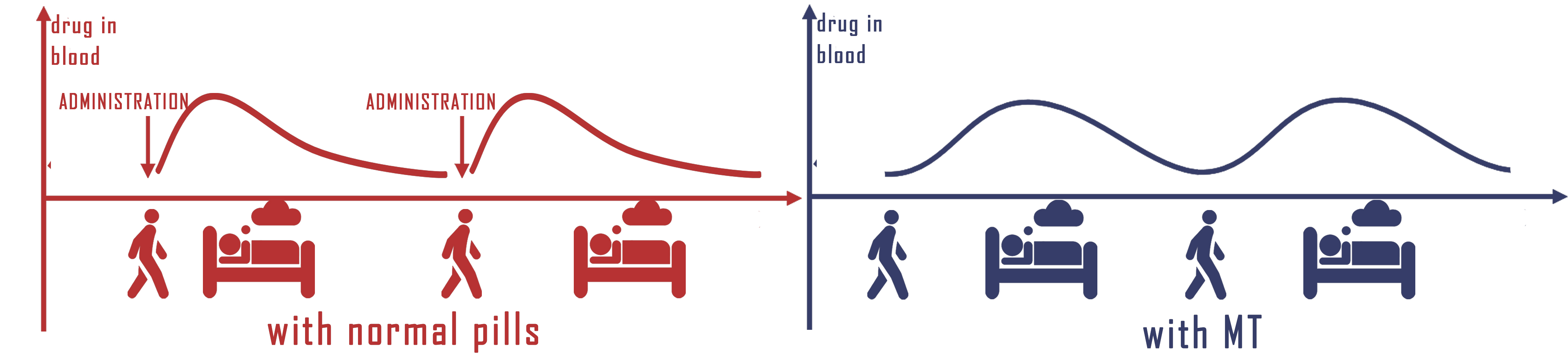
| | + | <p>b. Advantages of adding microbes rhythm in therapy </p> |
| | + | <p>Frankly speaking, almost everyone has the experience of not taking medicines on time. In fact, patient compliance has always been a significant problem effecting the therapeutic results. It is common that a patient has to take different pills in different time each day, using the engineered E.coli to secrete medicines with rhythm would greatly simplify the medications and improve drug therapy compliance, therefore, ensures the curative effect. What's more, long-termed treatments would become available for any diseases or symptoms associated with time.</p> |
| | + | |
| | + | <!--在图片标签外 套 如下标签,href=图片地址,可以点击放大图片 --> |
| | + | <a class="fancybox" href="https://static.igem.org/mediawiki/2015/7/74/LdwBG2.jpeg"> <!--- 就是这个 --> |
| | + | <img alt="" src="https://static.igem.org/mediawiki/2015/7/74/LdwBG2.jpeg"> |
| | + | </a><!--别忘了闭合--> |
| | + | |
| | + | |
| | + | |
| | + | <br> |
| | + | <br> |
| | + | <p>c. Meaning of hereditary time limits</p> |
| | + | <p>By far, many iGEM projects are based on the construction of strains used in medical treatments, but planting living microorganisms into human body has great potential safety hazards. However, if the microbes have time limit devices triggered by inverted modules which would let microbes commit suicide in calculated time, we might be able to use them to treat human. Such microorganisms would die naturally after function, and since the condition of inverted modules is hereditary, the suicide process of the group would not be disturbed by cell fission, which would avoid most of the potential hazards. Time limits are also capable of more sophisticated applications, for example, timed initiation of gene expression. In the future biological industries and biomedical fields, its prospects are almost limitless.</p> |
| | + | |
| | </div> | | </div> |
| | | | |
| − | <div id="Ideas-of-Timer" class="scrollto">
| + | |
| − | <h2>Ideas of Timer</h2>
| + | |
| − | <p>The Picture is not available</p>
| + | |
| − | </div>
| + | |
| − | | + | |
| | <div id="Recombinase-System" class="scrollto"> | | <div id="Recombinase-System" class="scrollto"> |
| | <h2>Recombinase System</h2> | | <h2>Recombinase System</h2> |
| − | <p>The Picture is not available</p>
| + | <p>The design of our project is mainly based on recombinase system, a complete recombinase system comprises of a kind of recombinase and its corresponding recombination target sites(RTSs). Here are the recombinase system's principle. |
| | + | <a href="https://static.igem.org/mediawiki/2015/thumb/4/49/LDW-REC.jpeg/728px-LDW-REC.jpeg" class="fancybox"><img src="https://static.igem.org/mediawiki/2015/thumb/4/49/LDW-REC.jpeg/728px-LDW-REC.jpeg" alt="a. when two RTSs are in the same direction, the sequences between will be cut out and form a circular DNA under the performances of corresponding recombinase. b. when two RTSs are in reverse direction, the sequences between will be flipped under the performances of corresponding recombinase."></a> |
| | + | <p class="figure">a. when two RTSs are in the same direction, the sequences between will be cut out and form a circular DNA under the performances of corresponding recombinase. b. when two RTSs are in reverse direction, the sequences between will be flipped under the performances of corresponding recombinase.</p> |
| | </div> | | </div> |
| | </div> | | </div> |
| | | | |
| − | <div id="Abstract" class="scrollto"> | + | |
| − | <h1>Abstract</h1>
| + | <div id="Description" class="scrollto"> |
| − | <p>When we talk about internal clock, we actually refer to the life cycle of higher animals and plants. People naturally get up when the sun rises and fall asleep as night comes. Thanks to our internal clock, we are able to do appropriate things at appropriate times. This year, we develop a plug-in for microbes, a system that can build up a living clock for microbes and guide them to live regularly as well as to work efficiently. Also, microbes with the system inserted acquire the concept of life span so that some dangerous bacterias will come to death in time before they cause any biohazard. Inverted components which based on recombinase system are introduced in our system. One inverted component is able to calculate a period of time, while several inverted components comprise of one time cycle. When we combine several time cycles together, a sequence with a timer’s function has been successfully formed. Through altering recombinases and their specific sites to different matches, we can get distinctive time cycles. We believe that the system will be of great importance to biological devices with time-related concepts.</p>
| + | <h1>Description</h1> |
| − | </div>
| + | |
| − | <div id="Overview" class="scrollto"> | + | |
| − | <h1>Overview</h1> | + | |
| | <div id="Matching-and-Testing" class="scrollto"> | | <div id="Matching-and-Testing" class="scrollto"> |
| | <h2>Matching and Testing</h2> | | <h2>Matching and Testing</h2> |
| − | <h3>Introduction of purpose</h3>
| + | <p>The goal of our group is to provide a systematic solution to Micro-timer construction, which includes optimization of synthetic elements and measurement of invertase module timing length. Hence, we developed a real-time invertase dynamics testing system to observe the performance of each combination of different elements. The real-time invertase dynamics testing system consists of two different plasmids in E. coli, namely pInv-gen and pInv-rep. pInv-gen generates invertase-EGFP fusion protein when induced; pInv-rep produces mcherry signal when inverted by invertase-EGFP. The dynamic pattern of the two signal can be mathematically analyzed, and the interval between bursts of green and red signals indicate the timing length of the invertase module. Furthermore, we tested effects of promoter, fusion sites, and ssra-tag in this system, and these elements are extremely helpful to enhance the performance of the device. With this system, we successfully tested the activity of invertases including Cre, Vika, Scre, Vcre, Dre, and Flp.</p> |
| − | <p>The basic idea of Micro-time system is to separate a long-period timing into small invertase device modules, and through appropriate combination of them, we can obtain a wide range of aimed time length for users to choose. However, for both E. coli and yeast, a successful timer must be based on precise definition and measurement of “time unit” – how long each invertase module exactly represents. Hence, the major consideration of our testing group is to measure the time unit for different invertase modules, and provide a systematic solution with optimized synthetic elements to gain a Micro-timer for any length of time (see Fig-T1).</p>
| + | </div> |
| | | | |
| − | <img alt="fig-1" src="">
| |
| | | | |
| − | <p>This mission require us to utilize a method to quantitatively understand the in vivo enzymatic dynamics of each invertase, and the system we conduct such experiment must be reconstruction-friendly, since we have to test a variety of elements (e.g. promoter and ssra) in similar pattern to most accurately obtain data. Hence, we developed a real-time invertase dynamics testing system to observe the performance of each combination of different elements.</p>
| |
| | | | |
| − | <h3>System construction</h3>
| + | <div id="Prokaryotic-Timer" class="scrollto"> |
| − | <p>The real-time invertase dynamics testing system contains two different plasmids in E. coli (see Fig-T2). The first one is an invertase generation vector, namely pInv-gen, that produces invertase-EGFP fusion protein through induction. The second one is called pInv-rep, a reporter vector that produce mcherry signal to indicate the inversion successfully happens. The invertase-EGFP on pInv-gen is controlled by an inducible promoter (T7-LacO promoter or Pbad). The target sequence (RTS) of corresponding invertase locates in the pInv-rep, surrounding a mcherry gene which is yet upside-down and transcribed by a constructive promoter (e.g. BBa_J23101). This mcherry-coding sequence can be inverted and restored to 5’ – 3’ direction at the existence of Cre-GFP, rendering red signal. Additionally, an ssra tag that intensifies the protein degradation may be fused to the C-terminus of invertase-EGFP and mcherry to be in tune with our final device that aims to clean up the redundant invertase not participating in a second round inversion.</p>
| + | <h2>Prokaryotic Timer</h2> |
| − | <img src="" alt="fig-2">
| + | <p>Our group aims at constructing an innovative timer in bacterial system based on recombinase-triggered flipping, in which we attempted to construct several parts and devices served for relevant function. ECFP, mCherry and GFP were detected in different time scale to exhibit wholly flipping process. We developed a creative joining method called "2A" assembly to cope with plentiful scattered DNA fragments, when, at last, we managed to get our circuits tested by certain ways on its availability.</p> |
| | | | |
| − | <p>Once if the inducer is added into the culture, the green fluorescence will increase at first due to the expression of invertase-EGFP. Then, the red fluorescence is generated because the Cre-EGFP restores the reversed mcherry sequence (see Fig-T-3). The length of interval between green and red indicates the corresponding single timing length of the invertase module. In our study, the variants to render different time length are invertase itself, promoter, and the degrading rate by ssra. Specifically, the activity level of invertase directly determines the time need to invert most of pInv-reps, and the promoter decides the rate of generation of invertase, which also contribute sigfificant to the speed of module. The ssra-tag, on the contrary, reduces the speed of inversion while effectively inhibiting the leakage expression when inducer is not in the culture.</p>
| |
| − | <img src="" alt="fig-3">
| |
| − |
| |
| − | <h3>Achievement</h3>
| |
| − | <p>We uses this system to measure totally 21 pairs of different combination of pInv-gens and pInv-reps. There are 6 different invertases we have tested using the Real-time system. While Cre and Flp are most commonly used recombinases in Biobrick plates, we newly contributed 4 brand-new invertases: Dre, Vcre, Scre, and Vika, all of which are Cre-family recombinases with different and non-intervolving RTS. We successfully proved that all these invertase work pretty good in our system, which you can see in <a href="">RESULTS</a>. All of the data we gather are analyzed by modeling group to render its corresponding time length. This work could guide other groups for their final design.</p>
| |
| − |
| |
| − | <h3>Timer design plug-in</h3>
| |
| − | <p>Additionally, we prepared a website plug-in to for potential users to design their own timer with specific length of counting time, a project in cooperation with SYSU-software. According to the data gathered in this research and other promoter intensity given by iGEM official page, we can anticipate the overall timing length of any given combination of various invertase module. Vice versus, if a user could provide his/her target time, we can automatically generate one or more optimized design of Micro-time to precisely match the demand.</p>
| |
| | </div> | | </div> |
| − | <div id="Bacteria-Timer" class="scrollto">
| |
| − | <h2>Bacteria Timer</h2>
| |
| − | <h3>Introduction</h3>
| |
| | | | |
| − | <p>A report from Science[1](在本版面最后添加参考文献), by which we were inspired, tries to explain that synthetic gene networks can be constructed to emulate a cellular counter that would enable complex synthetic programming and a variety of biotechnology applications.</p>
| |
| − | <p>One of the figures (fig. 1) from this article indicates how genes can work in a counting system by reversal of recombinases. Two recombinases in the circuit, Flpe and Cre, in conjunction with their specific recognition site, FRT and LoxP, accomplishes the whole flipping process. Convenient to distinguish, we'll call it circuit 1.</p>
| |
| − | <img src="" alt="fig-1-1">
| |
| | | | |
| − |
| |
| − | <p>Based on Circuit 1, we designed what we call circuit 2 (fig. 2) , which is, to our perspective, more functional and less induced, by principally altering the location of genes.</p>
| |
| − | <img src="" alt="fig-1-2">
| |
| − |
| |
| − | <p>The comparison between them indicates 2 advantages of circuit 2 over circuit 1 (fig. 3) . </p>
| |
| − | <p>First, it continues transcripting and flipping in a circulation once induced. It happens theoratically because it may be bothered by objective resistence, but it provides us with a possibility to time gene reaction and control certain protein expression in a time scale. </p>
| |
| − | <p>Second, circuit 2 can transfer certain DNA sequences unit after unit. Imagine if there is a target gene between the first frt and loxP in the initial phase, it would pass on and on in the continuous units after flipping.</p>
| |
| − | <img src="" alt="fig-1-3">
| |
| | | | |
| − |
| + | <div id="Telomeric-Timer" class="scrollto"> |
| − | <h3>Construction</h3>
| + | <h2>Telomeric Timer</h2> |
| | + | <p>Telomeric timer,also called Micro-timer 2.0, is designed to endue microorganisms with programed life span based on the cell cycle specific promoter (in eukaryotes) )or other cell division related promoter (in prokaryotes). In this system, the flanking RTSs are of same direction, enabling the sequences between them to be excised by specific recombinases, causing deletion of DNA fragments. With every cell division, this device will sequentially truncate a part of the sequence and finally lead to cell death when there is no more sequences to be truncated, working like a telomere.</p> |
| | + | </div> |
| | | | |
| − | <p>Due to time limits, we focused on constructing circuit 2, our original design.</p>
| + | <div id="Eukaryotic-Timer" class="scrollto"> |
| − | <p>We added florescent protein eCFP(标上砖号) and mCherry(标上砖号) right in the downstream of the ssrA-tag of recombinase flpe and cre, respectively. And we add a final GFP(标上砖号) as a reporter. pSB1C3 was used as vector for cloning and we tried to transfer the entire circuit to pSB3K3 in order to test its viability (fig. 4) .</p>
| + | <h2>Eukaryotic Timer</h2> |
| − | <img src="" alt="fig-1-4">
| + | <p>Eukaryotic timer,also called Micro-timer 3.0, uses recombinases from Ser family such as Bxb1, which typically catalyze site-specific recombination between an attachment site on the infecting phage chromosome (attP) and an attachment site in the host chromosome (attB) in natural system. The resulting integration reaction inserts the phage genome into the host chromosome bracketed by newly formed attL and attR (LR) sites. When attB and attP are engineered to be opposite BP sites, the integrase catalyzes the inversion of sequences flanked by BP sites, changing BP sites into LR sites, and will not revert the DNA flanked by LR sites. </p> |
| − |
| + | |
| − | <p>In terms of ligation efficiency, we resembles small fragments (promoters, FRTs and LoxPs) and deliver them to IDT for complete synthesis. Then we ligate long fragments in between according to our design (fig. 5) . Worth of attention, we created a new "2A" assembly by using DNA clean up and arranging gene segments with different resistances (fig. 6).</p>
| + | <p>In the design of Micro-timer 3.0, each inverted promoter flanked by BP sites is at the downstream of an inverted reporter gene and followed by a ser integrase gene. The reporter i(inverted reporter gene)-attP-promoter i-attB-integrase unit is defined as a counting motif, named eukaryotic timer integrase motif (EIM). EIM can be inserted into different sites of chromosomes, creating a large scale and multi-level system in eukaryotes.</p> |
| − | <img src="" alt="fig-1-5">
| + | |
| − | <img src="" alt="fig-1-6">
| + | <p>The circuit can be programmed to record time by counting a specific type of events such as the expression of cyclins. Once the motifs are activated, the downstream expression can work automatically and will not be terminated or reset by the hosts themselves, which is the reason why we believe that such a system can imprint “the same time” on microbes derived from a monoclone.</p> |
| − |
| + | |
| − | <h3>Testing</h3>
| + | <p>To make better use of the bxb1 integrase, we improved the biobrick BBa_K1039003 submitted by iGEM2013-Waterloo, optimizing it to match RFC 25 standard (BBa_K1641900) and thus it can be applied on 3A-fusion-assembly. |
| | | | |
| − | <p>Experimental proof must be accomplished after construction. </p>
| |
| − | <p>2 objectives for circuit 2 must be achieved during testing. One is that we must prove it actually has capability to reverse. The other one is that we must test its efficiency and explore how close it is to an ideal biological device that can really calculate time.</p>
| |
| − | <p>For the first goal, we thought that it could be solved by digestion (fig. 7) . When sequences flip, certain restricted enzyme cutting sites won’t change. What have changed are their locations. Gene length can be altered when sequences reverse, which can be visualized in an elecrophoretic way, as shown in fig. 7.</p>
| |
| − | <img src="" alt="fig-1-7">
| |
| − |
| |
| − | <p>For the second goal, we used qPCR to verify feasibility of the circuit. We design primers shown in the picture (fig. 8) . We can tell from amplication curves (fig. 9) whether it reverses and calculates time when phases have altered.</p>
| |
| − | <img src="" alt="fig-1-8">
| |
| − | <img src="" alt="fig-1-9">
| |
| − | </div>
| |
| − | <div id="Yeast-Timer" class="scrollto">
| |
| − | <h2>Yeast Timer</h2>
| |
| − | <p>(Our project, micro-timer, is to construct a counter on DNA that can imprint time on microbes.)</p>
| |
| − | <p>Micro timer 2.0 (Eu-timer) is constructed by DNA-based counting motifs that are inserted into different sites of chromosomes, creating a relatively large-scale system with more motifs than that in Micro timer 1.0.</p>
| |
| − |
| |
| − | <p>The Eu-timer uses recombinases from Ser family such as Bxb1, which typically catalyzes site-specific recombination between an attachment site on the infecting phage chromosome (attP) and an attachment site in the host chromosome (attB) in natural system. The resulting integration reaction inserts the phage genome into the host chromosome bracketed by newly formed attL and attR (LR) sites. When attB and attP are engineered to be opposite BP sites, the integrase alone catalyzes the inversion of sequences flanked by BP sites, changing BP sites into LR sites, and will not revert the DNA flanked by LR sites. </p>
| |
| − |
| |
| − | <p>In the design of Eu-timer (Fig 1) , each inverted promoter flanked by BP sites is downstream of an inverted reporter gene and followed by a ser integrase gene. The reporter i(inverted reporter gene)-attP-promoter i-attB-integrase unit is defined as a counting motif, named eu-timer integrase motif (EIM).</p>
| |
| − |
| |
| − | <p>The circuit can be programmed to record time by counting a specific type of events like the expression of cyclins. Once the motifs are activated, the downstream expression can work automatically and will not be terminated or reset by the hosts themselves, which is the reason why we believe that such a system can imprint “the same time” on microbes derived from a single clone.</p>
| |
| − |
| |
| − | <p>We then designed an telomere-like device by making little changes upon Micro timer 1.0, named Micro timer 1.1 (Fig 2) , in which the flanking site are of same direction. With every cell division, this device will sequentially truncate a part of the sequence, and finally lead to cell death, working like telomere.</p>
| |
| | </div> | | </div> |
| − | <div id="Modelling" class="scrollto">
| |
| − | <h2>Modelling</h2>
| |
| − | <p>First of all, we get two tables of one certain combination, including different kinds of plasmid with certain type of promoter, invert-ase together with its recognition site on the reporter and a ssra tag of a specific intensity which we will calculate later. From the second table in each group, we show the RFU changing with respect to time and obviously it reflects the quantity of the protein. The second derivative of the fitting curve is the enzymatic activity, which is the product of the enzymatic activity of one single enzyme at a given moment and the total quantity of the enzyme. Now let consider it separately. As for the enzymatic activity, we use the Michaelis-Menten equation to describe it. </p>
| |
| | | | |
| | | | |
| − | <p>S in this equation is the quantity of the plasmid to be inverted in the bacterial population per volume . S= S initial value-the first order integral of V0. So the expression of V0 is an ODE model. Another important fact that we need to take into consideration is that since the quantity of the plasmid to be inverted in the bacterial population per volume is limited, the total feedback is on the enzymatic activity. So the ODE model can be only used from the start moment till the first derivative of the curve reaching its maximum. As for the quantity of the enzyme, we can use the cftool in matlab to fit a function to show the quantity with respect to the time using the data in the first table of each group. However, in order to get the total quantity of the enzyme produced in the process we must add the expression leakage of RFU (expression: OD0/OD * G0 . OD0 is the first line of the first table in moment 0. OD is shown in table 3. G0 is also constant. ) to the actual value of the RFU in the first table of each group which supposed to be linear. Up to now, we can get the coefficient Vmax and Km in this function, that is, we get the exact function of the fitting curve of the second table in each group. In order to get the rest part of the function, let’s move forward to the meaning of the first order derivative of the fitting curve = quantity of inverted promoters equal (=S) * promoter intensity – the degeneration rate of the protein with the ssra tag – the nutrition correction equation which called Logistic equation. The degeneration rate of the protein with the ssra tag can be obtain in the following method. We use the principal component analysis on each four difference between row OCG and OCGS, OGC and OGCS, MCG and MCGS, MGC and MGCS of the same line which means at the same moment, to integrate one data for each line. Then we use the data at moment 0,1,2 and so on to construct the function of the degeneration rate of the protein caused by the ssra tag using the cftool in matlab. </p>
| |
| | | | |
| − | <p>Now we get the complete function of the second table and the constant Vmax and Km, which is the property of the certain kind of enzyme we use in each group.</p>
| + | <div id="Registry-Contribution" class="scrollto"> |
| − |
| + | <h2>Registry Contribution</h2> |
| − | <p>The abbreviation of the term in each row.</p>
| + | <h3>New application of Cre (BBa_K1179058) and its detailed measurement.</h3> |
| | + | <p>Although the data in the iGEM Registry is pretty large for iGEMers to obtain most common information, there is limited material about recombinases, and less about the invertase activity. Hence we tried our best to gather information about one of the most commonly used recombinases – Cre. </p> |
| | + | <p>BBa_K1179058 is a Cre CDS provided by team iGEM13_MIT, and all of our experiments about cre is established using this sequence. We developed a real-time invertase dynamics testing system based on BBa_K1179058, and confirmed its functionality as an invertase. Further, we measured the dynamics pattern of its enzymatic activity and discovered some interesting property of this invertase.</p> |
| | + | <p>In sum, we not only provided a new application for this part, but also measured the detailed dynamics pattern of this recombinase. </p> |
| | + | |
| | + | <h3>RFC25 optimization of bxb1integrase (BBa_K1039003)</h3> |
| | + | <p>The bxb1 integrase part has been submitted by iGEM2013-Waterloo (BBa_K1039003), but it does not match the RFC25 standard and thus can not be applied on fusion via 3A assembly. In SYSU-CHINA 2015, we modified the bxb1 gene to make it match the RFC25 standard.</p> |
| | + | |
| | | | |
| − | <p>Now we use the data of MCG group as an example, the fitting curve of the second table is the red one, the blue one is the scatter diagram linked together with line showing quantity of the enzyme with respect to the time and since we know the form of the function is linear, we can easily derivate the exact function by fitting.</p>
| |
| | </div> | | </div> |
| | </div> | | </div> |
| | | | |
| − | <div id="Collaboration" class="scrollto"> | + | |
| − | <h1>Collaboration</h1>
| + | |
| − | <div class="scrollto" id="Collaboration-with-ZJU">
| + | |
| − | <h2>Collaboration with ZJU</h2>
| + | |
| − | <p>This year, we cooperated with Zhejiang University in microbial suicide regulation. The project of ZTU this year will use microbial toxins in termite control and prevention, though how to improve strain safety become a important subject. However, one important part of our project is delayed suicide of microorganisms, so, in the hope of improving safety of their experiments, we provide them a complete regulatory suicide circuit, along which we also provide the detailed data of promotor's inducing expression intensity.</p>
| + | |
| − | </div>
| + | |
| − | <div class="scrollto" id="Collaboration-with-SJTU">
| + | |
| − | <h2>Collaboration with SJTU</h2>
| + | |
| − | <p>This year, SJTU_Software build a biobrick selection assist system based on empirical algorithm. This system can generalize the heat map of different splicing pattern, based on the history record of its splicing sequence in the database, and provide a experiential score to the user. Experiential score is a score that not associated to certain experiment, so a specific situation is needed, which means it grades the results under different experiment condition.</p>
| + | |
| − | <p>We cooperate with SJTU_Software on the project of experiment grading and result predicting. We would set up multiple parallel experiment groups in the construction and testing of inverted components. These groups are comparable between their designs and results, which would become one of the pivotal parts of our project, and the data extracted from which would meet SJTU_Software’s needs.</p>
| + | |
| − | <p>Through our cooperation, they can grade our experiment results, then the scores would be compared to our evaluation. To SJTU_Software, the comparison of our experiment results and the design of our plasmids' construction is a case which can directly evaluate their software. And it is also meaningful for us to evaluate the circuits with software, which would be beneficial to improve our future experiments.</p>
| + | |
| − | </div>
| + | |
| − | <div class="scrollto" id="Collaboration-with-THU">
| + | |
| − | <h2>Collaboration with THU</h2>
| + | |
| − | <p>
| + | |
| − |
| + | |
| − | </p>
| + | |
| − | </div>
| + | |
| − | </div>
| + | |
| | </div> | | </div> |
| | | | |
| | </html> | | </html> |
| | {{SYSU_CHINA_CLOSE}} | | {{SYSU_CHINA_CLOSE}} |