Difference between revisions of "Team:Waterloo/Lab Overview"
(Created page with "{{Waterloo}} <html> <div id ="mainContainer"> <div id ="contentContainer"> </div> </div> </html> {{Waterloo_Footer}}") |
(add cas9mod info) |
||
| (7 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
{{Waterloo}} | {{Waterloo}} | ||
<html> | <html> | ||
| − | <div | + | <div class="container main-container"> |
| − | + | ||
| − | </ | + | <h1>Wet Lab Overview</h1> |
| + | <p> | ||
| + | This summer Waterloo iGEM worked on making the CRISPR system more flexible and expand its applications. The wet lab focused in three aspects of this. A design of the modified sgRNA have been proposed in order to add restriction sites to exchange the target using classical cloning. In addition, Waterloo iGEM attempted to mutate dCas9 to recognize different PAM sites. Furthermore, we expanded the CRISPR-Cas9 system’s applications for eukaryotic systems, using <em>Arabodopsis thaliana</em> as the model organism. For more details on the design visit the links below. | ||
| + | </p> | ||
| + | |||
| + | <section id="sgRNA" title="sgRNA Modification" class="sgRNAmod"> | ||
| + | <h2><a href="https://2015.igem.org/Team:Waterloo/Lab/sgRNA">sgRNA Modification</a></h2> | ||
| + | <p> | ||
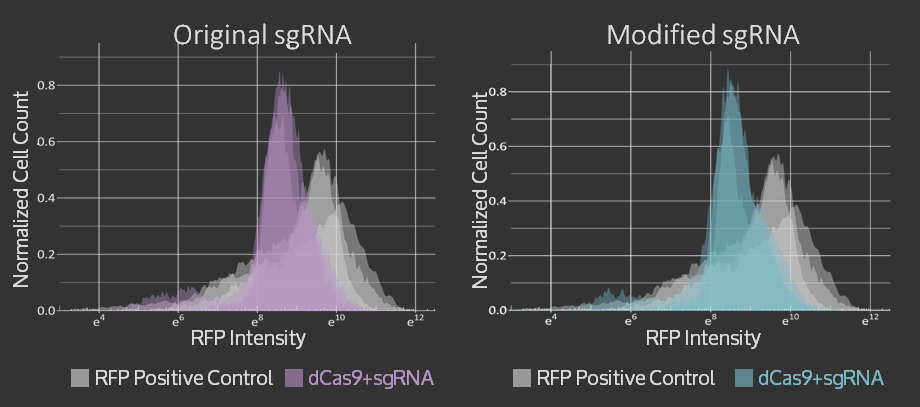
| + | <p>For the Simple sgRNA Exchange, we were able to show that adding restriction sites and a single mutation in the scaffold region of the sgRNA <strong>did not reduce targeting</strong> of the sgRNA in CRISPR dCas9 system. The graphs of flow cytometry data can be seen below and more details on the results are available on the <a href="https://2015.igem.org/Team:Waterloo/Lab/sgRNA">sgRNA Exchange page</a>. | ||
| + | </p> | ||
| + | <figure> | ||
| + | <img src="/wiki/images/0/09/Waterloo_flowcytometryresults.png" alt="CRISPR-Cas9 structure and project overview" /> | ||
| + | <figcaption>RFP Intensity after targeting with dCas9 + standard sgRNA and dCas9 + modified sgRNA in blue. Non-targeted RFP control measurements are shown in grey. Replicate measurements taken on three separate days are overlayed on the same axes.</figcaption> | ||
| + | </figure> | ||
| + | </p> | ||
| + | </section> | ||
| + | |||
| + | <section id="cas9" title="Cas9 Modification" class="cas9mod"> | ||
| + | <h2><a href="/Team:Waterloo/Lab/dCas9">Cas9 Modification</a></h2> | ||
| + | <p> | ||
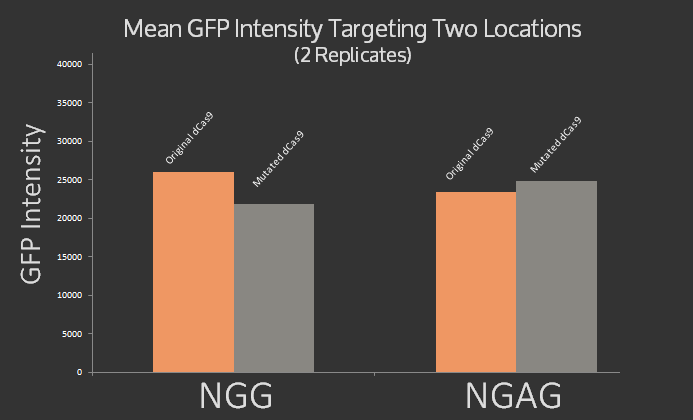
| + | In addition, we \mutated dCas9 at three sites 1135, 1335, and 1337 to alter its PAM site from native NGG to NGAG using the Quick Change protocol. The mutations were confirmed using sequencing. Preliminary experiments were done using sgRNA targeting the J23101 promoter upstream of the GFP using NGAG PAM site, however the results, graphed below, were inconclusive. It was expected that the original dCas9 would target NGG but not NGAG, while the modified dCas9 would target NGAG but not NGG. Instead roughly the same signal was measured for both dCas9 variants at both target sites. | ||
| + | </p> | ||
| + | |||
| + | <figure> | ||
| + | <img src="/wiki/images/9/95/Waterloo_pamflexlabresults.png" alt="Bar graphs showing results from dCas9 targeting" /> | ||
| + | <figcaption>Mean RFP Intensity after targeting an NGAG and NGG PAM site (using different complementary sgRNA sequences) with both dCas9 and the modified dCas9 variant.</figcaption> | ||
| + | </figure> | ||
| + | </section> | ||
| + | |||
| + | <section id="plants" title="Plant Defence" class="plantmod"> | ||
| + | <h2>Plant Viral Defence</h2> | ||
| + | <p> | ||
| + | <a href="/Team:Waterloo/Lab/Plants">Plant Defence</a> | ||
| + | </p> | ||
| + | </section> | ||
</div> | </div> | ||
</html> | </html> | ||
{{Waterloo_Footer}} | {{Waterloo_Footer}} | ||
Latest revision as of 23:39, 20 November 2015
Wet Lab Overview
This summer Waterloo iGEM worked on making the CRISPR system more flexible and expand its applications. The wet lab focused in three aspects of this. A design of the modified sgRNA have been proposed in order to add restriction sites to exchange the target using classical cloning. In addition, Waterloo iGEM attempted to mutate dCas9 to recognize different PAM sites. Furthermore, we expanded the CRISPR-Cas9 system’s applications for eukaryotic systems, using Arabodopsis thaliana as the model organism. For more details on the design visit the links below.
sgRNA Modification
For the Simple sgRNA Exchange, we were able to show that adding restriction sites and a single mutation in the scaffold region of the sgRNA did not reduce targeting of the sgRNA in CRISPR dCas9 system. The graphs of flow cytometry data can be seen below and more details on the results are available on the sgRNA Exchange page.
Cas9 Modification
In addition, we \mutated dCas9 at three sites 1135, 1335, and 1337 to alter its PAM site from native NGG to NGAG using the Quick Change protocol. The mutations were confirmed using sequencing. Preliminary experiments were done using sgRNA targeting the J23101 promoter upstream of the GFP using NGAG PAM site, however the results, graphed below, were inconclusive. It was expected that the original dCas9 would target NGG but not NGAG, while the modified dCas9 would target NGAG but not NGG. Instead roughly the same signal was measured for both dCas9 variants at both target sites.