Difference between revisions of "Team:Bielefeld-CeBiTec/Results/HeavyMetals"
| Line 42: | Line 42: | ||
<h1>Arsenic</h1> | <h1>Arsenic</h1> | ||
<h2><i>in vivo</i></h2> | <h2><i>in vivo</i></h2> | ||
| + | <p>We tested an arsenic sensor with mRFP1 as reporter gene <i>in vivo</i> to confirm that the sensor is functional and test whether it is possible to detect the safety limit as defined by the WHO. We observed a reaction approximately five hours after addition of arsenic. The safety limit of 10 µg/L could clearly be distinguished from the negative control and the fluorescence signal increased up to a concentration of 500 µg/L. The signal in the presence of 1000 µg/L was slightly lower than in the presence of 500 µg/L.</p> | ||
<figure style="width: 600px"> | <figure style="width: 600px"> | ||
| − | <a href="https://static.igem.org/mediawiki/2015/ | + | <a href="https://static.igem.org/mediawiki/2015/9/9d/Bielefeld_CeBiTec_arsenic_invivo_large.png" data-lightbox="heavymetals" data-title="Time course of the induction of an arsenic biosensor with RFP for different arsenic concentrations <i>in vivo</i>. Error bars represent the standard deviation of three biological replicates."><img src="https://static.igem.org/mediawiki/2015/6/63/Bielefeld_CeBiTec_arsenic_invivo_small.png" alt="Adjusting the detection limit"></a> |
<figcaption>Time course of the induction of an arsenic biosensor with RFP for different arsenic concentrations in vivo. Error bars represent the standard deviation of three biological replicates.</figcaption> | <figcaption>Time course of the induction of an arsenic biosensor with RFP for different arsenic concentrations in vivo. Error bars represent the standard deviation of three biological replicates.</figcaption> | ||
</figure> | </figure> | ||
<h2><i>in vitro</i></h2> | <h2><i>in vitro</i></h2> | ||
| + | <p>In order to test the arsenic sensor in our cell-free protein synthesis, we cloned a device that contains the arsenic operator between the T7 promoter and sfGFP with our optimized untranslated region (UTR). We tested this device in a cell extract that had been generated from cells expressing the arsenic repressor. We observed an induction when adding arsenic up to a concentration of 1.87 mg/L. As high arsenic concentrations inhibit the performance of the CFPS, we normalized the results for this effect. In the final application, this task is performed by our app.</p> | ||
<figure style="width: 600px"> | <figure style="width: 600px"> | ||
| − | <a href="https://static.igem.org/mediawiki/2015/1/19/Bielefeld_CeBiTec_arsenic_corrected_large.png" data-lightbox="heavymetals" data-title="Induction of arsenic sensor in vitro. For this experiment, a cell extract which already containes the arsenic repressor was used. Error bars represent the standard deviation of three biological replicates."><img src="https://static.igem.org/mediawiki/2015/1/14/Bielefeld_CeBiTec_arsenic_corrected_small.png" alt="Adjusting the detection limit"></a> | + | <a href="https://static.igem.org/mediawiki/2015/1/19/Bielefeld_CeBiTec_arsenic_corrected_large.png" data-lightbox="heavymetals" data-title="Induction of arsenic sensor <i>in vitro</i>. For this experiment, a cell extract which already containes the arsenic repressor was used. Error bars represent the standard deviation of three biological replicates."><img src="https://static.igem.org/mediawiki/2015/1/14/Bielefeld_CeBiTec_arsenic_corrected_small.png" alt="Adjusting the detection limit"></a> |
<figcaption>Induction of arsenic sensor in vitro. For this experiment, a cell extract which already containes the arsenic repressor was used. Error bars represent the standard deviation of three biological replicates.</figcaption> | <figcaption>Induction of arsenic sensor in vitro. For this experiment, a cell extract which already containes the arsenic repressor was used. Error bars represent the standard deviation of three biological replicates.</figcaption> | ||
</figure> | </figure> | ||
Revision as of 13:47, 14 September 2015
Heavy Metals
Zusammenfassung in ganz wenigen Worten.

Arsenic
in vivo
We tested an arsenic sensor with mRFP1 as reporter gene in vivo to confirm that the sensor is functional and test whether it is possible to detect the safety limit as defined by the WHO. We observed a reaction approximately five hours after addition of arsenic. The safety limit of 10 µg/L could clearly be distinguished from the negative control and the fluorescence signal increased up to a concentration of 500 µg/L. The signal in the presence of 1000 µg/L was slightly lower than in the presence of 500 µg/L.

in vitro
In order to test the arsenic sensor in our cell-free protein synthesis, we cloned a device that contains the arsenic operator between the T7 promoter and sfGFP with our optimized untranslated region (UTR). We tested this device in a cell extract that had been generated from cells expressing the arsenic repressor. We observed an induction when adding arsenic up to a concentration of 1.87 mg/L. As high arsenic concentrations inhibit the performance of the CFPS, we normalized the results for this effect. In the final application, this task is performed by our app.

Chromium
in vivo
Our sensor for chromium detection consists of ChrB the repressor and the chromate specific promoter ChrP. The promoter is regulated by the ChrB, which binds Cr-ions. Behind the promoter is a sfGFP for detection of a fluorescence signal. In vivo we could show that the addition of different concentrations of chromium have different effects to transcription of sfGFP.

in vitro





Lead
in vivo


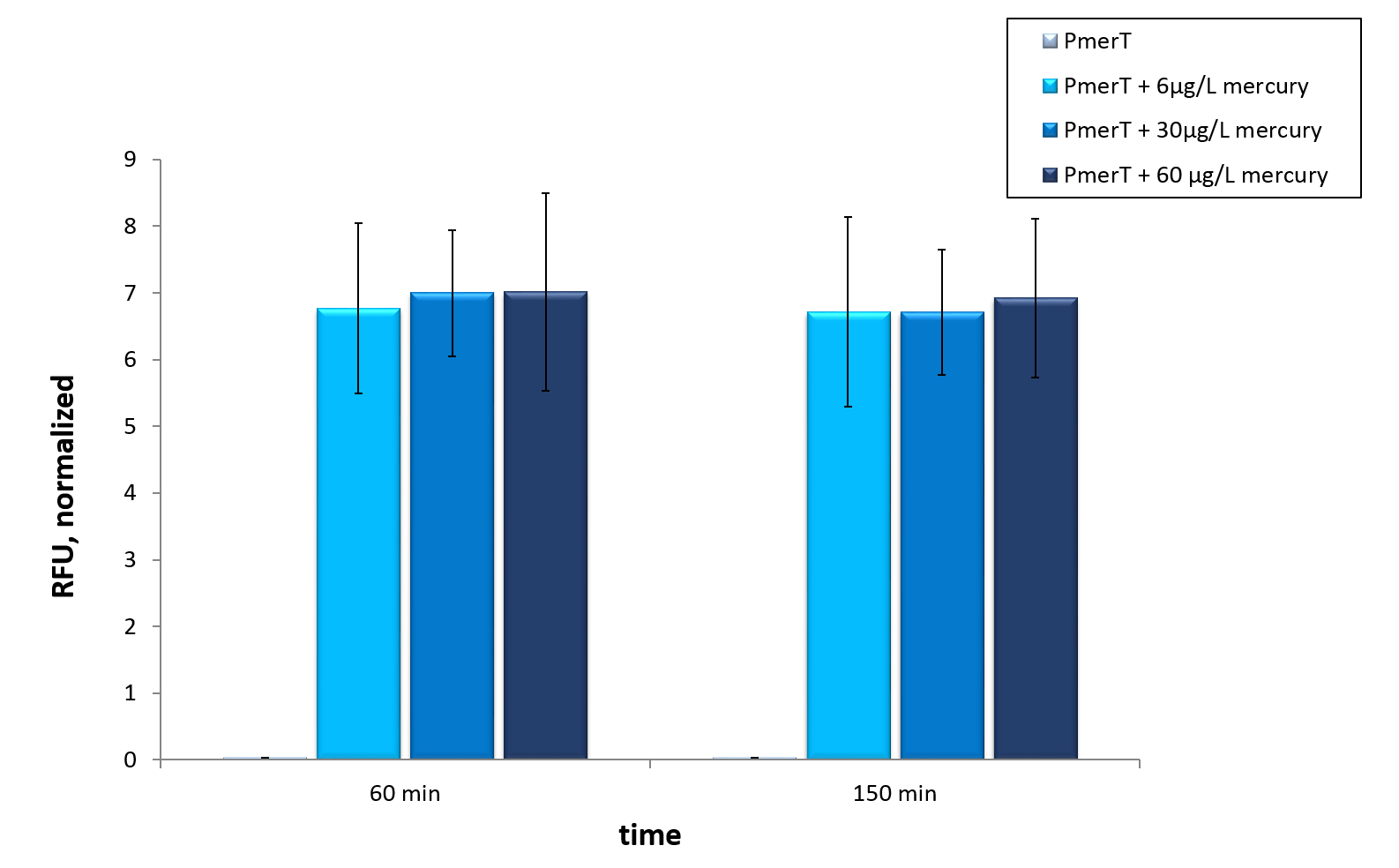
Mercury
in vivo


in vitro



Nickel
in vivo



