Difference between revisions of "Team:Bielefeld-CeBiTec/Results/HeavyMetals"
| Line 226: | Line 226: | ||
<h2><i>in vivo</i></h2> | <h2><i>in vivo</i></h2> | ||
| − | <p>The chromium sensor (<a href="http://parts.igem.org/Part:BBa_K1758313" target="_blank">BBa_K1758313</a>) was constructed by using the basic construction we showed in <a href="https://2015.igem.org/Team:Bielefeld-CeBiTec/Project/HeavyMetals" target="_blank">Our biosensors</a>. We work with the chromate inducible operon of <i>Ochrobactrum tritici</i> 5bvl1 which enables a resistance for chromium VI and superoxide. For our Sensor we used the Cr (VI) dependent repressor chrB which was introduced by team BIT 2013 (<a href="http://parts.igem.org/Part:BBa_K1758313" target="_blank">BBa_K1058007</a>), and optimized this sequence for the use in <EM> E. coli </EM>. The associated chromium responsive promoter is ChrP (introduced by BIT 2013 (<a href="http://parts.igem.org/Part:BBa_K1758313" target="_blank">BBa_K1058007</a> | + | <p>The chromium sensor (<a href="http://parts.igem.org/Part:BBa_K1758313" target="_blank">BBa_K1758313</a>) was constructed by using the basic construction we showed in <a href="https://2015.igem.org/Team:Bielefeld-CeBiTec/Project/HeavyMetals" target="_blank">Our biosensors</a>. We work with the chromate inducible operon of <i>Ochrobactrum tritici</i> 5bvl1 which enables a resistance for chromium VI and superoxide. For our Sensor we used the Cr (VI) dependent repressor chrB which was introduced by team BIT 2013 (<a href="http://parts.igem.org/Part:BBa_K1758313" target="_blank">BBa_K1058007</a>), and optimized this sequence for the use in <EM> E. coli </EM>. The associated chromium responsive promoter is ChrP (introduced by BIT 2013 (<a href="http://parts.igem.org/Part:BBa_K1758313" target="_blank">BBa_K1058007</a>). For output we used sfGFP and a 5’UTR untranslated region in front of sfGFP to optimize the expression of the reporter protein and increase its fluorescence.</p> |
| Line 307: | Line 307: | ||
<figure style="width: 600px"> | <figure style="width: 600px"> | ||
<a href="https://static.igem.org/mediawiki/2015/b/b2/Bielefeld-CebiTec_in_vivo_Copper.jpeg" data-lightbox="heavymetals" data-title=" "><img src="https://static.igem.org/mediawiki/2015/b/b2/Bielefeld-CebiTec_in_vivo_Copper.jpeg" alt="genetical approach"></a> | <a href="https://static.igem.org/mediawiki/2015/b/b2/Bielefeld-CebiTec_in_vivo_Copper.jpeg" data-lightbox="heavymetals" data-title=" "><img src="https://static.igem.org/mediawiki/2015/b/b2/Bielefeld-CebiTec_in_vivo_Copper.jpeg" alt="genetical approach"></a> | ||
| − | <figcaption>Construct konst.Prom + CueR+CopAP-UTR-sfGFP <a href="http://parts.igem.org/Part: BBa_K1758324" target="_blank"> BBa_K1758324</a> consisting of konst.Prom + CueR <a href="http://parts.igem.org/Part: BBa_K1758320" target="_blank">BBa_K17583240</a> and CopAP-UTR-sfGF <a href="http://parts.igem.org/Part:BBa_K1758323" target="_blank">BBa_K1758323</a> used for<i>in vivo</i> characterization.</figcaption> | + | <figcaption>Construct konst.Prom + CueR+CopAP-UTR-sfGFP <a href="http://parts.igem.org/Part:BBa_K1758324" target="_blank">BBa_K1758324</a> consisting of konst.Prom + CueR <a href="http://parts.igem.org/Part:BBa_K1758320" target="_blank">BBa_K17583240</a> and CopAP-UTR-sfGF <a href="http://parts.igem.org/Part:BBa_K1758323" target="_blank">BBa_K1758323</a> used for<i>in vivo</i> characterization.</figcaption> |
</figure> | </figure> | ||
| Line 327: | Line 327: | ||
<h2><i>in vitro</i></h2> | <h2><i>in vitro</i></h2> | ||
| − | <p>For the characterization of the copper sensor with CFPS we used parts differing from that we used in vivo characterization. For the in vitro characterization we used a cell extract out of cells which contain the Plasmid (<a href="http://parts.igem.org/Part: BBa_K1758320" target="_blank"> BBa_K1758320</a>). In addition to that we added Plasmid-DNA of the copper specific promoter copAP with 5’UTR-sfGFP under the control of T7-promoter (<a href="http://parts.igem.org/Part: BBa_K1758324" target="_blank"> BBa_K1758324</a>)to the cell extract. The T7-promoter is needed to get a better fluorescence expression. </p> | + | <p>For the characterization of the copper sensor with CFPS we used parts differing from that we used in vivo characterization. For the in vitro characterization we used a cell extract out of cells which contain the Plasmid (<a href="http://parts.igem.org/Part:BBa_K1758320" target="_blank">BBa_K1758320</a>). In addition to that we added Plasmid-DNA of the copper specific promoter copAP with 5’UTR-sfGFP under the control of T7-promoter (<a href="http://parts.igem.org/Part:BBa_K1758324" target="_blank">BBa_K1758324</a>)to the cell extract. The T7-promoter is needed to get a better fluorescence expression. </p> |
<figure style="width: 600px"> | <figure style="width: 600px"> | ||
<a href="https://static.igem.org/mediawiki/2015/0/05/Bielefeld-CeBiTec_in_vitro_CueR-part.jpeg" data-title=" "><img src="https://static.igem.org/mediawiki/2015/0/05/Bielefeld-CeBiTec_in_vitro_CueR-part.jpeg"></a> | <a href="https://static.igem.org/mediawiki/2015/0/05/Bielefeld-CeBiTec_in_vitro_CueR-part.jpeg" data-title=" "><img src="https://static.igem.org/mediawiki/2015/0/05/Bielefeld-CeBiTec_in_vitro_CueR-part.jpeg"></a> | ||
| − | <figcaption> konst.Prom + CueR <a href="http://parts.igem.org/Part: BBa_K1758320" target="_blank"> BBa_K1758320</a> used for<i>in vitro</i> characterization.</figcaption> | + | <figcaption> konst.Prom + CueR <a href="http://parts.igem.org/Part:BBa_K1758320" target="_blank">BBa_K1758320</a> used for<i>in vitro</i> characterization.</figcaption> |
</figure> | </figure> | ||
<figure style="width: 600px"> | <figure style="width: 600px"> | ||
<a href="https://static.igem.org/mediawiki/2015/1/15/Bielefeld-CebiTec_in_vitro_T7-copAP-UTR-sfGFP.jpeg" data-lightbox="heavymetals" data-title=" "><img src="https://static.igem.org/mediawiki/2015/1/15/Bielefeld-CebiTec_in_vitro_T7-copAP-UTR-sfGFP.jpeg"></a> | <a href="https://static.igem.org/mediawiki/2015/1/15/Bielefeld-CebiTec_in_vitro_T7-copAP-UTR-sfGFP.jpeg" data-lightbox="heavymetals" data-title=" "><img src="https://static.igem.org/mediawiki/2015/1/15/Bielefeld-CebiTec_in_vitro_T7-copAP-UTR-sfGFP.jpeg"></a> | ||
| − | <figcaption> T7-copAP-UTR-sfGFP <a href="http://parts.igem.org/Part: BBa_K1758324" target="_blank"> BBa_K1758324</a> used for<i>in vitro</i> characterization.</figcaption> | + | <figcaption> T7-copAP-UTR-sfGFP <a href="http://parts.igem.org/Part:BBa_K1758324" target="_blank">BBa_K1758324</a> used for<i>in vitro</i> characterization.</figcaption> |
</figure> | </figure> | ||
| Line 394: | Line 394: | ||
<p>In addition to these we constructed a sensor for lead detection. It consists of PbrR, the repressor, and the lead specific promoter PbrA. The promoter is regulated by the RcnR, which binds Pb-ions. As the former sensors this one encloses a sfGFP for detection via fluorescence. </p> | <p>In addition to these we constructed a sensor for lead detection. It consists of PbrR, the repressor, and the lead specific promoter PbrA. The promoter is regulated by the RcnR, which binds Pb-ions. As the former sensors this one encloses a sfGFP for detection via fluorescence. </p> | ||
| − | <p>Our lead sensor consists of parts of the chromosomal lead operon of <EM> Cupriavidusmetallidurans (Ralstoniametallidurans) </EM>. This operon includes the promoter PbrA (<a href="http://parts.igem.org/Part:BBa_K1758332" target="_blank"> BBa_K1758332 </a>) , which is regulated by the repressor pbrR. The PbrR belongs to the MerR family, of metal-sensing regulatoryproteins, and is Pb2+-inducible. Our sensor system comprises PbrR (<a href="http://parts.igem.org/Part:BBa_K1758330" target="_blank"> BBa_K1758330 </a>), which is under the control of a constitutive Promoter and PbrA and a 5’ untranslated region, which controls the transcription of a sfGFP and increases the fluorescence. Fluorescence implemented by sfGFP protein is the measured output signal. </p> | + | <p>Our lead sensor consists of parts of the chromosomal lead operon of <EM> Cupriavidusmetallidurans (Ralstoniametallidurans) </EM>. This operon includes the promoter PbrA (<a href="http://parts.igem.org/Part:BBa_K1758332" target="_blank">BBa_K1758332 </a>) , which is regulated by the repressor pbrR. The PbrR belongs to the MerR family, of metal-sensing regulatoryproteins, and is Pb2+-inducible. Our sensor system comprises PbrR (<a href="http://parts.igem.org/Part:BBa_K1758330" target="_blank"> BBa_K1758330 </a>), which is under the control of a constitutive Promoter and PbrA and a 5’ untranslated region, which controls the transcription of a sfGFP and increases the fluorescence. Fluorescence implemented by sfGFP protein is the measured output signal. </p> |
<figure style="width: 600px"> | <figure style="width: 600px"> | ||
<a href="https://static.igem.org/mediawiki/2015/a/a3/Bielefeld-CebiTec_in_vivo_Lead.jpeg" data-lightbox="heavymetals" data-title=" "><img src="https://static.igem.org/mediawiki/2015/a/a3/Bielefeld-CebiTec_in_vivo_Lead.jpeg" alt="genetical approach"></a> | <a href="https://static.igem.org/mediawiki/2015/a/a3/Bielefeld-CebiTec_in_vivo_Lead.jpeg" data-lightbox="heavymetals" data-title=" "><img src="https://static.igem.org/mediawiki/2015/a/a3/Bielefeld-CebiTec_in_vivo_Lead.jpeg" alt="genetical approach"></a> | ||
| − | <figcaption>Construct konst.Prom + PbrR+CopAP-UTR-sfGFP <a href="http://parts.igem.org/Part: BBa_K1758334" target="_blank"> BBa_K1758334</a> consisting of konst.Prom + PbrR <a href="http://parts.igem.org/Part: BBa_K1758330" target="_blank"> BBa_K17583230</a> and PbrA-UTR-sfGF <a href="http://parts.igem.org/Part: BBa_K1758333" target="_blank"> BBa_K1758333</a> used for<i>in vivo</i> characterization.</figcaption> | + | <figcaption>Construct konst.Prom + PbrR+CopAP-UTR-sfGFP <a href="http://parts.igem.org/Part:BBa_K1758334" target="_blank"> BBa_K1758334</a> consisting of konst.Prom + PbrR <a href="http://parts.igem.org/Part:BBa_K1758330" target="_blank"> BBa_K17583230</a> and PbrA-UTR-sfGF <a href="http://parts.igem.org/Part:BBa_K1758333" target="_blank"> BBa_K1758333</a> used for<i>in vivo</i> characterization.</figcaption> |
</figure> | </figure> | ||
| Line 435: | Line 435: | ||
<figure style="width: 600px"> | <figure style="width: 600px"> | ||
<a href="https://static.igem.org/mediawiki/2015/0/0d/Bielefeld-CebiTec_in_vivo_Mercury.jpeg" data-lightbox="heavymetals" data-title=" "><img src="https://static.igem.org/mediawiki/2015/0/0d/Bielefeld-CebiTec_in_vivo_Mercury.jpeg"></a> | <a href="https://static.igem.org/mediawiki/2015/0/0d/Bielefeld-CebiTec_in_vivo_Mercury.jpeg" data-lightbox="heavymetals" data-title=" "><img src="https://static.igem.org/mediawiki/2015/0/0d/Bielefeld-CebiTec_in_vivo_Mercury.jpeg"></a> | ||
| − | <figcaption>Construct konst.Prom + MerR+MerT-UTR-sfGFP <a href="http://parts.igem.org/Part: BBa_K1758344" target="_blank"> BBa_K1758344</a> consisting of konst.Prom + MerR <a href="http://parts.igem.org/Part: BBa_K1758340" target="_blank"> BBa_K1758340</a> and MerT-UTR-sfGF <a href="http://parts.igem.org/Part: BBa_K1758342" target="_blank"> BBa_K1758342</a> used for<i>in vivo</i> characterization.</figcaption> | + | <figcaption>Construct konst.Prom + MerR+MerT-UTR-sfGFP <a href="http://parts.igem.org/Part:BBa_K1758344" target="_blank"> BBa_K1758344</a> consisting of konst.Prom + MerR <a href="http://parts.igem.org/Part:BBa_K1758340" target="_blank"> BBa_K1758340</a> and MerT-UTR-sfGF <a href="http://parts.igem.org/Part:BBa_K1758342" target="_blank"> BBa_K1758342</a> used for<i>in vivo</i> characterization.</figcaption> |
</figure> | </figure> | ||
| Line 456: | Line 456: | ||
<h2><i>in vitro</i></h2> | <h2><i>in vitro</i></h2> | ||
| − | <p>For the characterization of the mercury sensor with CFPS we used parts differing from that we used in vivo characterization. For the in vitro characterization we used a cell extract out of cells which contain the Plasmid (<a href="http://parts.igem.org/Part: BBa_K1758340" target="_blank"> BBa_K1758340</a>). In addition to that we added Plasmid-DNA of the copper specific promoter merT with 5’UTR-sfGFP under the control of T7-promoter (<a href="http://parts.igem.org/Part: BBa_K1758344" target="_blank"> BBa_K1758344</a>)to the cell extract. The T7-promoter is needed to get a better fluorescence expression. </p> | + | <p>For the characterization of the mercury sensor with CFPS we used parts differing from that we used in vivo characterization. For the in vitro characterization we used a cell extract out of cells which contain the Plasmid (<a href="http://parts.igem.org/Part:BBa_K1758340" target="_blank"> BBa_K1758340</a>). In addition to that we added Plasmid-DNA of the copper specific promoter merT with 5’UTR-sfGFP under the control of T7-promoter (<a href="http://parts.igem.org/Part:BBa_K1758344" target="_blank"> BBa_K1758344</a>)to the cell extract. The T7-promoter is needed to get a better fluorescence expression. </p> |
<figure style="width: 600px"> | <figure style="width: 600px"> | ||
<a href="https://static.igem.org/mediawiki/2015/3/3c/Bielefeld-CeBiTec_in_vitro_merR-part.jpeg" data-title=" "><img src="https://static.igem.org/mediawiki/2015/3/3c/Bielefeld-CeBiTec_in_vitro_merR-part.jpeg"></a> | <a href="https://static.igem.org/mediawiki/2015/3/3c/Bielefeld-CeBiTec_in_vitro_merR-part.jpeg" data-title=" "><img src="https://static.igem.org/mediawiki/2015/3/3c/Bielefeld-CeBiTec_in_vitro_merR-part.jpeg"></a> | ||
| − | <figcaption> konst.Prom + MerR <a href="http://parts.igem.org/Part: BBa_K175840" target="_blank"> BBa_K1758340</a> used for<i>in vitro</i> characterization.</figcaption> | + | <figcaption> konst.Prom + MerR <a href="http://parts.igem.org/Part:BBa_K175840" target="_blank"> BBa_K1758340</a> used for<i>in vitro</i> characterization.</figcaption> |
</figure> | </figure> | ||
<figure style="width: 600px"> | <figure style="width: 600px"> | ||
<a href="https://static.igem.org/mediawiki/2015/e/e2/Bielefeld-CebiTec_in_vitro_T7-merT-UTR-sfGFP.jpeg" data-lightbox="heavymetals" data-title=" "><img src="https://static.igem.org/mediawiki/2015/e/e2/Bielefeld-CebiTec_in_vitro_T7-merT-UTR-sfGFP.jpeg"></a> | <a href="https://static.igem.org/mediawiki/2015/e/e2/Bielefeld-CebiTec_in_vitro_T7-merT-UTR-sfGFP.jpeg" data-lightbox="heavymetals" data-title=" "><img src="https://static.igem.org/mediawiki/2015/e/e2/Bielefeld-CebiTec_in_vitro_T7-merT-UTR-sfGFP.jpeg"></a> | ||
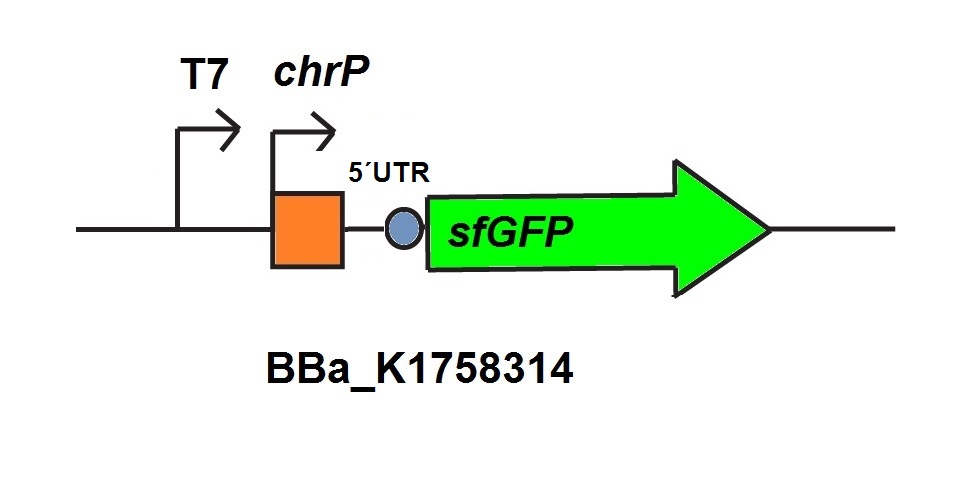
| − | <figcaption> T7-merT-UTR-sfGFP <a href="http://parts.igem.org/Part: BBa_K1758344" target="_blank"> BBa_K175834</a> used for<i>in vitro</i> characterization.</figcaption> | + | <figcaption> T7-merT-UTR-sfGFP <a href="http://parts.igem.org/Part:BBa_K1758344" target="_blank"> BBa_K175834</a> used for<i>in vitro</i> characterization.</figcaption> |
</figure> | </figure> | ||
| Line 510: | Line 510: | ||
<figure style="width: 600px"> | <figure style="width: 600px"> | ||
<a href="https://static.igem.org/mediawiki/2015/8/8e/Bielefeld-CebiTec_in_vivo_Nickel.jpeg" data-lightbox="heavymetals" data-title=" "><img src="https://static.igem.org/mediawiki/2015/8/8e/Bielefeld-CebiTec_in_vivo_Nickel.jpeg"></a> | <a href="https://static.igem.org/mediawiki/2015/8/8e/Bielefeld-CebiTec_in_vivo_Nickel.jpeg" data-lightbox="heavymetals" data-title=" "><img src="https://static.igem.org/mediawiki/2015/8/8e/Bielefeld-CebiTec_in_vivo_Nickel.jpeg"></a> | ||
| − | <figcaption>Construct konst.Prom + rcnR+rcnA-UTR-sfGFP <a href="http://parts.igem.org/Part: BBa_K1758354" target="_blank"> BBa_K1758354</a> consisting of konst.Prom + rcnR <a href="http://parts.igem.org/Part: BBa_K1758350" target="_blank"> BBa_K1758340</a> and rcnA-UTR-sfGF <a href="http://parts.igem.org/Part: BBa_K1758352" target="_blank"> BBa_K1758352</a> used for<i>in vivo</i> characterization.</figcaption> | + | <figcaption>Construct konst.Prom + rcnR+rcnA-UTR-sfGFP <a href="http://parts.igem.org/Part:BBa_K1758354" target="_blank"> BBa_K1758354</a> consisting of konst.Prom + rcnR <a href="http://parts.igem.org/Part:BBa_K1758350" target="_blank"> BBa_K1758340</a> and rcnA-UTR-sfGF <a href="http://parts.igem.org/Part:BBa_K1758352" target="_blank"> BBa_K1758352</a> used for<i>in vivo</i> characterization.</figcaption> |
</figure> | </figure> | ||
Revision as of 20:36, 16 September 2015
Heavy Metals
Zusammenfassung in ganz wenigen Worten.
The different sensors we worked with were characterized in vivo as well as in vitro.
We tested the influence of each heavy metal on our sensors in vivo Therefore we used heavy metal concentrations based on heavy metal occurrences measured all over the world.

The tested heavy metal concentrations had no negative effect on E. colis growth. Moreover there is no significant difference between the curves with heavy metals and the controls. This first experiment showed us, in vivo characterization with these sensors under the tested heavy metal concentrations is possible. Most of our sensors were cultivated in the BioLector. Due to the accuracy of this device we could measure our sample in duplicates.
Click on the test strip for more information about the heavy metals and how they can be detected: