Difference between revisions of "Team:Bielefeld-CeBiTec/Results/HeavyMetals"
| Line 240: | Line 240: | ||
| − | <p>Our sensor for chromium detection consists of ChrB the repressor and the chromate specific promoter ChrP. The promoter is regulated by ChrB, which binds Cr-ions. Behind the promoter is a sfGFP for detection of a fluorescence signal.</p> | + | <p>Our sensor for chromium detection consists of ChrB the repressor and the chromate specific promoter ChrP. The promoter is regulated by ChrB, which binds Cr<sup>6+</sup>-ions. Behind the promoter is a sfGFP for detection of a fluorescence signal.</p> |
<figure style="width: 600px"> | <figure style="width: 600px"> | ||
| Line 252: | Line 252: | ||
<div class="col-md-6 text-center" style="margin-bottom: 50px"> <figure style="width: 600px"> | <div class="col-md-6 text-center" style="margin-bottom: 50px"> <figure style="width: 600px"> | ||
<a href="https://static.igem.org/mediawiki/2015/8/82/Bielefeld-CeBiTec_Biolector_chromium.jpg" data-lightbox="heavymetals" data-title="Time course of the induction of a chromium biosensor with sfGFP for different chromium concentrations <i>in vivo</i>. The data are measured with BioLector and normalized on OD<sub>600</sub>. Error bars represent the standard deviation of two biological replicates. ."><img src="https://static.igem.org/mediawiki/2015/8/82/Bielefeld-CeBiTec_Biolector_chromium.jpg" alt="Adjusting the detection limit"></a> | <a href="https://static.igem.org/mediawiki/2015/8/82/Bielefeld-CeBiTec_Biolector_chromium.jpg" data-lightbox="heavymetals" data-title="Time course of the induction of a chromium biosensor with sfGFP for different chromium concentrations <i>in vivo</i>. The data are measured with BioLector and normalized on OD<sub>600</sub>. Error bars represent the standard deviation of two biological replicates. ."><img src="https://static.igem.org/mediawiki/2015/8/82/Bielefeld-CeBiTec_Biolector_chromium.jpg" alt="Adjusting the detection limit"></a> | ||
| − | <figcaption>Time course of the induction of a chromium biosensor with sfGFP for different chromium concentrations in vivo. The data are measured with BioLector and normalized on | + | <figcaption>Time course of the induction of a chromium biosensor with sfGFP for different chromium concentrations in vivo. The data are measured with BioLector and normalized on OD<sub>600</sub>. Error bars represent the standard deviation of two biological replicates.</figcaption> |
</figure> | </figure> | ||
</div> | </div> | ||
| Line 345: | Line 345: | ||
<h2><i>in vivo</i></h2> | <h2><i>in vivo</i></h2> | ||
| − | <p>Our sensor for copper detection consists of CueR a MerR like activator and the copper specific promoter CopAP. The promoter is regulated by CueR, which binds | + | <p>Our sensor for copper detection consists of CueR a MerR like activator and the copper specific promoter CopAP. The promoter is regulated by CueR, which binds Cu<sup>2+</sup>-ions. We also used a sfGFP behind the promoter for detection trough a fluorescence signal.</p> |
<p>For our copper sensor we used the native operator of cooper homeostasis from <i> E.coli </i> K12. And constructed a part (BBa_K1758324) which was constructed using the basic construction showed in <a href="https://2015.igem.org/Team:Bielefeld-CeBiTec/Project/HeavyMetals" target="_blank">Our biosensors</a>.This operator includes the promoter (copAP), which is regulated by the repressor CueR. This is an important part for the aerobic copper tolerance. In BBa_K1758324 we combined the codon optimized CueR (<a href="http://parts.igem.org/Part:BBa_K1758320" target="_blank">BBa_K1758320</a>) under the control of a constitutive promoter with CopAP and sfGFP (<a href="http://parts.igem.org/Part:BBa_K1758321" target="_blank">BBa_K1758321</a>) for measuring output signals. Through the addition of a 5’UTR before the sfGFP we optimized the expression of sfGFP and increased fluorescence. </p> | <p>For our copper sensor we used the native operator of cooper homeostasis from <i> E.coli </i> K12. And constructed a part (BBa_K1758324) which was constructed using the basic construction showed in <a href="https://2015.igem.org/Team:Bielefeld-CeBiTec/Project/HeavyMetals" target="_blank">Our biosensors</a>.This operator includes the promoter (copAP), which is regulated by the repressor CueR. This is an important part for the aerobic copper tolerance. In BBa_K1758324 we combined the codon optimized CueR (<a href="http://parts.igem.org/Part:BBa_K1758320" target="_blank">BBa_K1758320</a>) under the control of a constitutive promoter with CopAP and sfGFP (<a href="http://parts.igem.org/Part:BBa_K1758321" target="_blank">BBa_K1758321</a>) for measuring output signals. Through the addition of a 5’UTR before the sfGFP we optimized the expression of sfGFP and increased fluorescence. </p> | ||
| Line 359: | Line 359: | ||
<div class="col-md-6 text-center" style="margin-bottom: 50px"> | <div class="col-md-6 text-center" style="margin-bottom: 50px"> | ||
<figure style="width: 600px"> | <figure style="width: 600px"> | ||
| − | <a href="https://static.igem.org/mediawiki/2015/9/90/Bielefeld-CeBiTec_Biolector_copper.jpg" data-lightbox="heavymetals" data-title="Time course of the induction of a copper biosensor with sfGFP for different copper concentrations in vivo. The data are measured with BioLector and normalized on | + | <a href="https://static.igem.org/mediawiki/2015/9/90/Bielefeld-CeBiTec_Biolector_copper.jpg" data-lightbox="heavymetals" data-title="Time course of the induction of a copper biosensor with sfGFP for different copper concentrations in vivo. The data are measured with BioLector and normalized on OD<sub>600</sub>. Error bars represent the standard deviation of two biological replicates."><img src="https://static.igem.org/mediawiki/2015/9/90/Bielefeld-CeBiTec_Biolector_copper.jpg" alt="Adjusting the detection limit"></a> |
| − | <figcaption>Time course of the induction of a copper biosensor with sfGFP for different copper concentrations in vivo. The data are measured with BioLector and normalized on | + | <figcaption>Time course of the induction of a copper biosensor with sfGFP for different copper concentrations in vivo. The data are measured with BioLector and normalized on OD<sub>600</sub>. Error bars represent the standard deviation of two biological replicates.</figcaption> |
</figure> | </figure> | ||
</div> | </div> | ||
| Line 467: | Line 467: | ||
<h2><i>in vivo</i></h2></br> | <h2><i>in vivo</i></h2></br> | ||
| − | <p>In addition to these we constructed a sensor for lead detection. It consists of PbrR, the repressor, and the lead specific promoter PbrAP. The promoter is regulated by the RcnR, which binds Pb-ions. As the former sensors this one encloses a sfGFP for detection via fluorescence. </p> | + | <p>In addition to these we constructed a sensor for lead detection. It consists of PbrR, the repressor, and the lead specific promoter PbrAP. The promoter is regulated by the RcnR, which binds Pb<sup>2+</sup>-ions. As the former sensors this one encloses a sfGFP for detection via fluorescence. </p> |
| − | <p>Our lead sensor consists of parts of the chromosomal lead operon of <EM> Cupriavidusmetallidurans (Ralstoniametallidurans) </EM>. This operon includes the promoter PbrAP (<a href="http://parts.igem.org/Part:BBa_K1758332" target="_blank">BBa_K1758332 </a>) , which is regulated by the repressor pbrR. The PbrR belongs to the MerR family, of metal-sensing regulatoryproteins, and is | + | <p>Our lead sensor consists of parts of the chromosomal lead operon of <EM> Cupriavidusmetallidurans (Ralstoniametallidurans) </EM>. This operon includes the promoter PbrAP (<a href="http://parts.igem.org/Part:BBa_K1758332" target="_blank">BBa_K1758332 </a>) , which is regulated by the repressor pbrR. The PbrR belongs to the MerR family, of metal-sensing regulatoryproteins, and is Pb<sup>2+</sup>-inducible. Our sensor system comprises PbrR (<a href="http://parts.igem.org/Part:BBa_K1758330" target="_blank"> BBa_K1758330 </a>), which is under the control of a constitutive Promoter and PbrAP and a 5’ untranslated region, which controls the transcription of a sfGFP and increases the fluorescence. Fluorescence implemented by sfGFP protein is the measured output signal. </p> |
<figure style="width: 600px"> | <figure style="width: 600px"> | ||
| Line 478: | Line 478: | ||
<div class="row"> | <div class="row"> | ||
<div class="col-md-6 text-center" style="margin-bottom: 50px"> <figure style="width: 600px"> | <div class="col-md-6 text-center" style="margin-bottom: 50px"> <figure style="width: 600px"> | ||
| − | <a href="http://https://static.igem.org/mediawiki/2015/d/d5/Bielefeld-CeBiTec_Biolector_lead.jpg" data-lightbox="heavymetals" data-title="Time course of the induction of a lead biosensor with sfGFP for different lead concentrations in vivo. The data are measured with BioLector and normalized on | + | <a href="http://https://static.igem.org/mediawiki/2015/d/d5/Bielefeld-CeBiTec_Biolector_lead.jpg" data-lightbox="heavymetals" data-title="Time course of the induction of a lead biosensor with sfGFP for different lead concentrations in vivo. The data are measured with BioLector and normalized on OD<sub>600</sub>. Error bars represent the standard deviation of two biological replicates. "><img src="https://static.igem.org/mediawiki/2015/d/d5/Bielefeld-CeBiTec_Biolector_lead.jpg" alt="Adjusting the detection limit"></a> |
| − | <figcaption>Time course of the induction of a lead biosensor with sfGFP for different lead concentrations in vivo. The data are measured with BioLector and normalized on | + | <figcaption>Time course of the induction of a lead biosensor with sfGFP for different lead concentrations in vivo. The data are measured with BioLector and normalized on OD<sub>600</sub>. Error bars represent the standard deviation of two biological replicates. </figcaption> |
</figure> | </figure> | ||
</div> | </div> | ||
| Line 510: | Line 510: | ||
Increased fluorescence signal after induction with mercury.--> | Increased fluorescence signal after induction with mercury.--> | ||
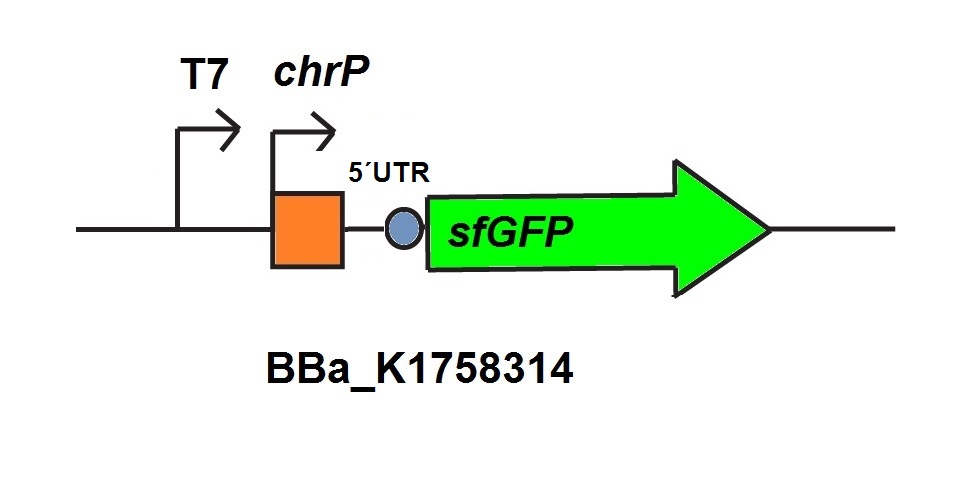
| − | <p>One of the already existing sensors we use for our system is the mercury sensor consisting of MerR the activator and the mercury specific promoter PmerT. The promoter is regulated by the MerR, which binds Hg-ions. Similar to the former sensors we added a sfGFP for detection via fluorescence. </p></br> | + | <p>One of the already existing sensors we use for our system is the mercury sensor consisting of MerR the activator and the mercury specific promoter PmerT. The promoter is regulated by the MerR, which binds Hg<sup>2+</sup>-ions. Similar to the former sensors we added a sfGFP for detection via fluorescence. </p></br> |
<p>For our mercury sensor we use parts of the mercury sensor constructed by iGEM team Peking 2010. The parts of iGEM team Peking 2010 consist of the mercury dependent Mer operon from <EM>shigella flexneri R100</EM> plasmid Tn21. The expression of the Mer operon is depends on the regulation by MerR its activator and promoter PmerT. For our sensor we used the codon optimized activator (<a href="http://parts.igem.org/Part:BBa_K1758340" target="_blank">BBa_K1758340</a>), under control of a constitutive promoter, of iGEM Peking 2010 (<a href="http://parts.igem.org/Part:BBa_K346001" target="_blank">BBa_K346001</a>). Additionally to the activator of Peking 2010 we used the specific promoter PmerT (<a href="http://parts.igem.org/Part:BBa_K346002" target="_blank">BBa_K346002</a>) from this team. For our sensor we added a 5’UTR behind this promoter to increase the fluorscence of the used reporter protein sfGFP.</p> | <p>For our mercury sensor we use parts of the mercury sensor constructed by iGEM team Peking 2010. The parts of iGEM team Peking 2010 consist of the mercury dependent Mer operon from <EM>shigella flexneri R100</EM> plasmid Tn21. The expression of the Mer operon is depends on the regulation by MerR its activator and promoter PmerT. For our sensor we used the codon optimized activator (<a href="http://parts.igem.org/Part:BBa_K1758340" target="_blank">BBa_K1758340</a>), under control of a constitutive promoter, of iGEM Peking 2010 (<a href="http://parts.igem.org/Part:BBa_K346001" target="_blank">BBa_K346001</a>). Additionally to the activator of Peking 2010 we used the specific promoter PmerT (<a href="http://parts.igem.org/Part:BBa_K346002" target="_blank">BBa_K346002</a>) from this team. For our sensor we added a 5’UTR behind this promoter to increase the fluorscence of the used reporter protein sfGFP.</p> | ||
| Line 544: | Line 544: | ||
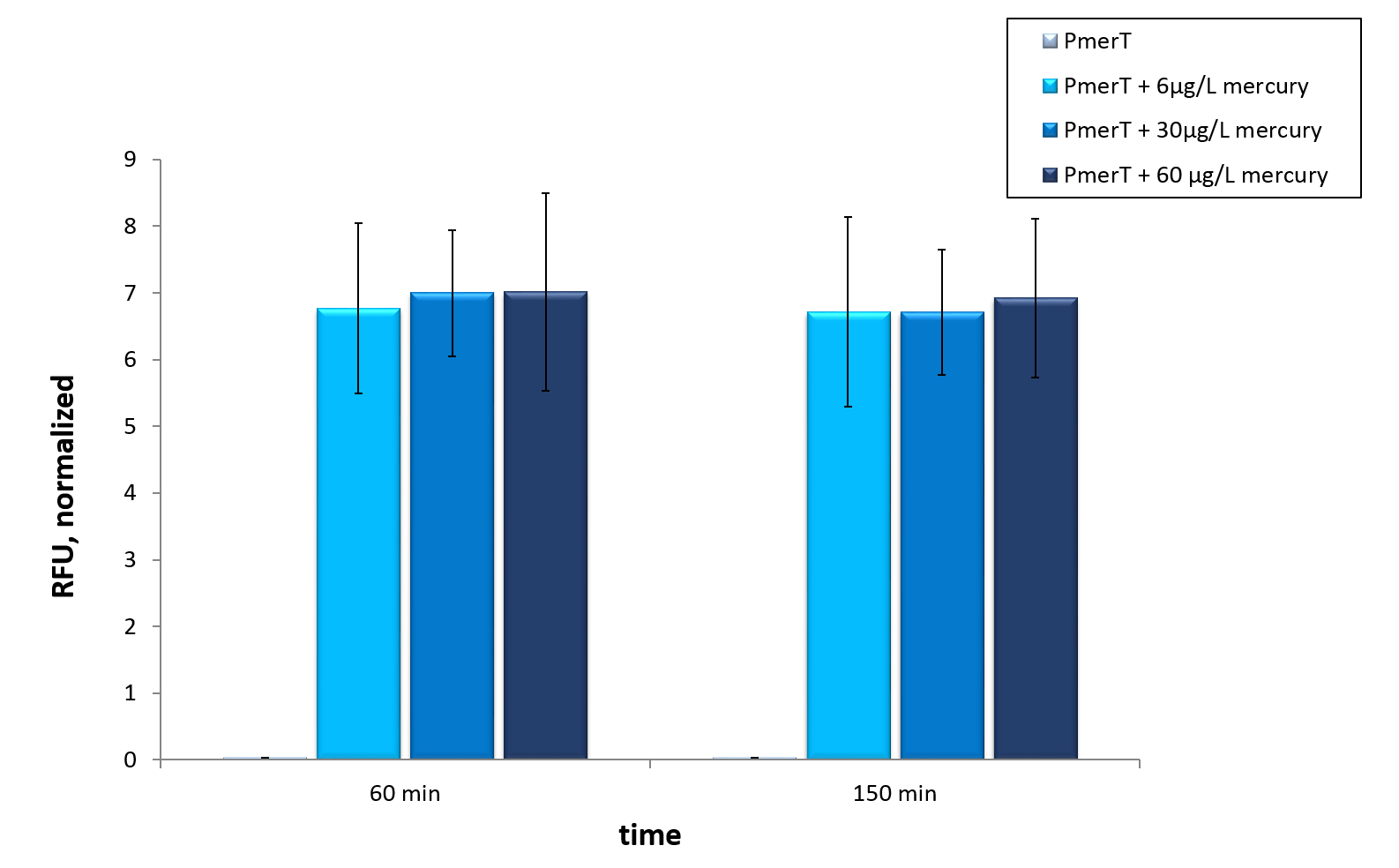
| − | <p>The mercury detection was measured during the cultivation of <i>E. coli</i> KRX at 37 °C. The strain contains the plasmid with the activator MerRunder the control of a constitutive promoter and the specific promoter with operator site which reacts to the activator with bound Hg-ions. The specific promoter is in front of sfGFP for measurment , so the mercury in the medium is detected directly.<i>In vivo</i> this sensor devise shows a fast answer to occurrence of his heavy metal contrary to the other sensor systems <i>in vivo</i>.</p> | + | <p>The mercury detection was measured during the cultivation of <i>E. coli</i> KRX at 37 °C. The strain contains the plasmid with the activator MerRunder the control of a constitutive promoter and the specific promoter with operator site which reacts to the activator with bound Hg<sup>2+</sup>-ions. The specific promoter is in front of sfGFP for measurment , so the mercury in the medium is detected directly.<i>In vivo</i> this sensor devise shows a fast answer to occurrence of his heavy metal contrary to the other sensor systems <i>in vivo</i>.</p> |
<p> Therefore we tested our sensor <i>in vitro</i> to check if an already functioning highly optimized sensor provides required data for guideline detections </p> | <p> Therefore we tested our sensor <i>in vitro</i> to check if an already functioning highly optimized sensor provides required data for guideline detections </p> | ||
| Line 605: | Line 605: | ||
<h1>Nickel</h1> | <h1>Nickel</h1> | ||
| − | <p> Naturally | + | <p> Naturally nickel occurrences are quite low. In Germany most drinking water pollutions by nickel happen in the last meters of the plumbing system. The reason for these pollutions are wrong tap ware (kreusche). There may be higher concentrations in drinking-water in special cases of release from natural or industrial nickel deposits. Higher nickel concentrations in water can lead to dermatitis as itching of fingers and other parts of the body through long term skin contact. The World Health Organization recommends a limit of 70 µg/L in drinking water. </p> |
<h2><i>in vivo</i></h2> | <h2><i>in vivo</i></h2> | ||
| − | <p>In addition to these we aimed to construct a sensor for nickel detection. It consists of RcnR the repressor and the nickel specific promoter PrcnA. The promoter is regulated by the RcnR, which binds Ni-ions. As the former sensors this one encloses a sfGFP for detection via | + | <p>In addition to these we aimed to construct a sensor for nickel detection. It consists of RcnR the repressor and the nickel specific promoter PrcnA. The promoter is regulated by the RcnR, which binds Ni<sup>2+</sup>-ions. As the former sensors this one encloses a sfGFP for detection via fluorescence.</p> |
| − | <p> Our | + | <p> Our nickel biosensor consists of parts of the rcn-operon from <i> E. coli </i> which codes for a nickel- and cobalt-efflux system. This system is highly sensitive to nickel. In absence of nickel or cobalt RcnR binds to the operator and inhibits the nickel responsive promoter. With Ni<sup>2+</sup>-ions present the repression of the promoter PrcnA will be reversed, because the repressor RcnR binds ni<sup>2+</sup>-ions and cannot attach to the DNA. For our biosensor we construct the part (<a href="http://parts.igem.org/Part:BBa_K1758353" target="_blank"> BBa_K1758353 </a>by using the basic construction showed in <Our biosensors >. For this part we used the repressor RcnR under control of a constitutive promoter (<a href="http://parts.igem.org/Part:BBa_K1758350" target="_blank"> BBa_K1758350 </a>) and the nickel specific promoter PrcnA with a 5’UTR in front of sfGFP (<a href="http://parts.igem.org/Part:BBa_K1758352" target="_blank"> BBa_K1758352 </a>) as reporter protein. </p> |
<figure style="width: 600px"> | <figure style="width: 600px"> | ||
| Line 620: | Line 620: | ||
<div class="row"> | <div class="row"> | ||
<div class="col-md-6 text-center" style="margin-bottom: 50px"><figure style="width: 600px"> | <div class="col-md-6 text-center" style="margin-bottom: 50px"><figure style="width: 600px"> | ||
| − | <a href="https://static.igem.org/mediawiki/2015/c/c9/Bielefeld-CeBiTec_Biolector_nickel.jpg" data-lightbox="heavymetals" data-title="Time course of the induction of a lead biosensor with sfGFP for different nickel concentrations in vivo. The data are measured with BioLector and normalized on | + | <a href="https://static.igem.org/mediawiki/2015/c/c9/Bielefeld-CeBiTec_Biolector_nickel.jpg" data-lightbox="heavymetals" data-title="Time course of the induction of a lead biosensor with sfGFP for different nickel concentrations in vivo. The data are measured with BioLector and normalized on OD<sub>600</sub>. Error bars represent the standard deviation of two biological replicates. "><img src="https://static.igem.org/mediawiki/2015/c/c9/Bielefeld-CeBiTec_Biolector_nickel.jpg" alt="Adjusting the detection limit"></a> |
| − | <figcaption>Time course of the induction of a lead biosensor with sfGFP for different nickel concentrations in vivo. The data are measured with BioLector and normalized on | + | <figcaption>Time course of the induction of a lead biosensor with sfGFP for different nickel concentrations in vivo. The data are measured with BioLector and normalized on OD<sub>600</sub>. Error bars represent the standard deviation of two biological replicates. </figcaption> |
</figure> | </figure> | ||
</div> | </div> | ||
| Line 632: | Line 632: | ||
<p> The data for our nickel sensor show a trend that differs for that of the other sensors. There is no indication for a working sensor <i>in vivo </i>. There is a fluorescence signal but it decreases in the first 5 hours. After reaching a minimum the fluorescence increases slowly. Additionally there is no difference in fluorescence as response to various nickel concentration. Nickels influence on the cells could be lead to a fallout which could result in decrease of fluorescence. | <p> The data for our nickel sensor show a trend that differs for that of the other sensors. There is no indication for a working sensor <i>in vivo </i>. There is a fluorescence signal but it decreases in the first 5 hours. After reaching a minimum the fluorescence increases slowly. Additionally there is no difference in fluorescence as response to various nickel concentration. Nickels influence on the cells could be lead to a fallout which could result in decrease of fluorescence. | ||
| − | With this sensor no production of | + | With this sensor no production of sfGFP via fluorescence level change could be detected. Therefore this sensor is not suitable for approach. Therefore no <i> in vitro </i> data using CFPS were taken.</p></br> |
<h2>To sum it up</h2> | <h2>To sum it up</h2> | ||
Revision as of 14:13, 18 September 2015
Heavy Metals
To make a long story short.

We tested our heavy metal biosensors in Escherichia coli as well as in our cell-free protein synthesis.
Prior to the in vivo characterization, we tested whether the heavy metals have a negative effect on the growth of E. coli.
As can be seen from the figure, we observed no significant difference between the growth in the presence of heavy metals and the controls. This first experiment showed us that in vivo characterization of these sensors is possible. Most cultivations for in vivo characterization were performed in the BioLector. Due to the accuracy of this device, we could measure our samples in duplicates. Subsequently, all functional biosensors were tested in vitro.
Click on the test strip for the results of our biosensor tests in E. coli and in our CFPS:







To sum it all up
We have characterized heavy metal sensors for arsenic, chromium, copper, lead, mercury and nickel. The results for our nickel characterization indicated that the constructed nickel sensor is not suitable for our test strip. The sensors for lead and chromium showed great potential, as they showed responses to chromium or lead, but require further optimization. Copper, our new heavy metal sensor, worked as expected and was able to detect different copper concentrations. The already well-characterized sensors for arsenic and mercury were tested as well. While the arsenic sensor worked well in vivo, it requires some omptimization for the use in vitro. Mercury showed that a fully optimized sensor works very well in our in vitro system and has the potential to detect even lower concentrations than in vivo.