Difference between revisions of "Team:Waterloo/Modeling"
| Line 9: | Line 9: | ||
</section> | </section> | ||
<section id="cas9_dynamics" title="Cas9 Frameshift Dynamics"> | <section id="cas9_dynamics" title="Cas9 Frameshift Dynamics"> | ||
| − | <h2>Cas9 Frameshift Dynamics</h2> | + | <h2>Cas9 Frameshift Dynamics</h2><div class="row"> |
| − | + | <div class="col-sm-8"> | |
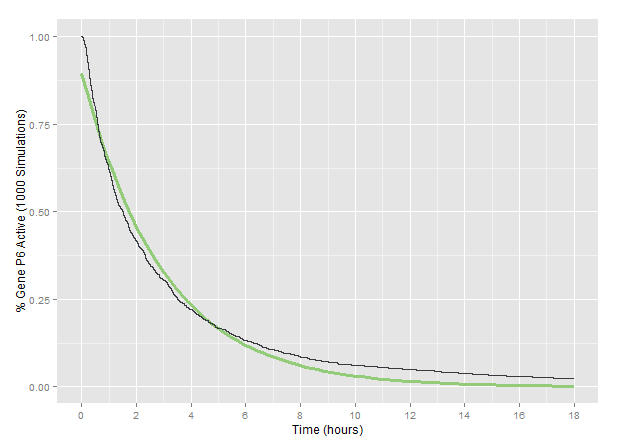
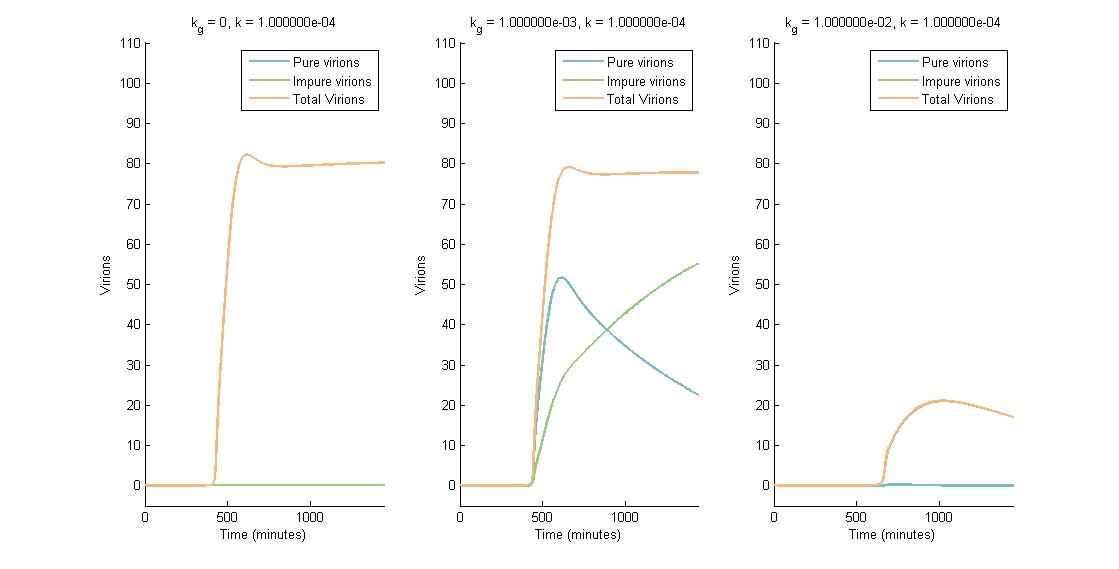
| + | We modelled the accumulation of mutations in a target genome and eventual deactivation of target genes after cutting by CRISPR/Cas9 and repair by Non-Homologous End Joining (NHEJ). A stochastic and probabilistic model of the effect of CRISPR/Cas9 on a single genome allowed us to investigate the accumulation of indels in a genome with multiple targets over time. Our Python model provides a tool for other teams looking to investigate multiplexed Cas9 targeting for deactivation of target genomes. | ||
| + | </div> | ||
| + | <div class="col-sm-4"> | ||
| + | <img src="https://static.igem.org/mediawiki/2015/1/15/Waterloo_camv_cut.png" class="img-responsive"/> | ||
| + | </div> | ||
| + | </div> | ||
| + | <figure> | ||
| + | <img src="wiki/images/c/c9/Waterloo_case2w1000.pngclass="img-responsive"/> | ||
| + | <figcaption>Image of agent-based model of spread of viral infection among plant cells sorted into several leaves.</figcaption> | ||
| + | </figure> | ||
</section> | </section> | ||
</section> | </section> | ||
Revision as of 03:59, 19 September 2015
Modeling
Mathematical modeling is a core part of Waterloo iGEM: we have nearly as many team members typing furiously away in our dry lab as we do wrangling transformations in our wet lab. This year, we created models in Python, MATLAB and NetLogo that examine the feasibility of our design and provide tools for assessing future designs. The code for each model is available on our GitHub and details on the formulation of each model may be found in the pages linked below.
Cas9 Frameshift Dynamics


PAM Structural Bioinformatics
A key limitation of the current use of S. pyogenes Cas9 for gene editing is the inability to target sequences which are not adjacent to a NGG PAM sequence. To overcome this issue, we developed a pipeline to probe Cas9 mutants with altered PAM specificity using simulation tools in PyRosetta and Python. A detailed description of the analysis and software used in this work can be found on the Modeling Engineered Cas9 PAM Flexibility.
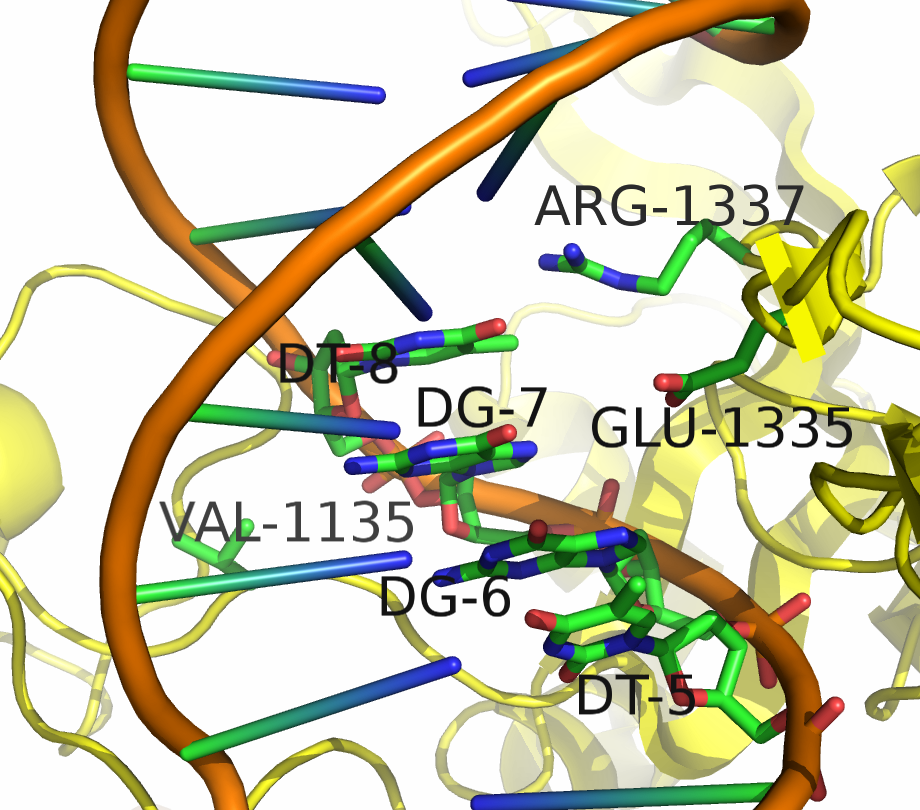
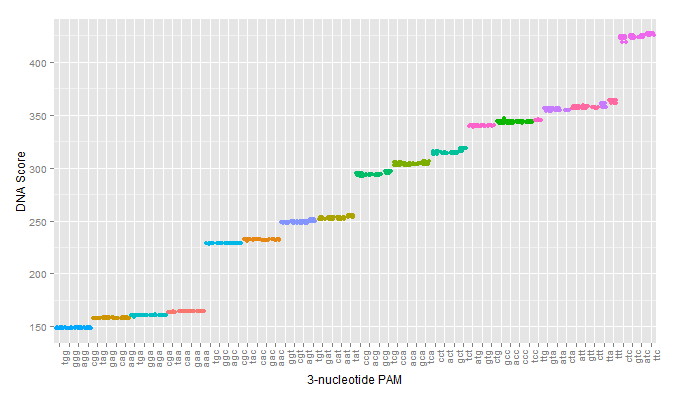
The simulation managed to successfully predict the PAM specificity of wild type spCAs9. However, validation against known Cas9 mutants was less successful, predicting a mix of correct and incorrect, though biochemically similar, PAM sequences.
Overall, our multi-scale model of CRISPR Plant Defense allowed us to investigate the extent to which we could slow CaMV spread using CRISPR/Cas9 and its interaction with host defense systems such as RNA interference and intercellular defense signalling.