Difference between revisions of "Team:DTU-Denmark"
| Line 221: | Line 221: | ||
<div class="container-fluid" id="scrollspy"> | <div class="container-fluid" id="scrollspy"> | ||
<ul class="nav navbar-nav"> | <ul class="nav navbar-nav"> | ||
| − | <li><a class="page-scroll" href="#Project- | + | <li><a class="page-scroll" href="#Project-Description">Project Description</a></li><li><a class="page-scroll" href="#Social-Media">Social Media</a></li> |
</ul> | </ul> | ||
</div> | </div> | ||
| Line 242: | Line 242: | ||
<li> | <li> | ||
| − | <a id="Project- | + | <a id="Project-Description-submenu" class="btn btn-default btn-transparent btn-lg page-scroll" href="#Project-Description">Project Description</a> |
</li> | </li> | ||
| Line 253: | Line 253: | ||
<li> | <li> | ||
| − | <a id="Project- | + | <a id="Project-Description-submenu" class="page-scroll" href="#Project-Description">Project Description</a> |
</li> | </li> | ||
| Line 266: | Line 266: | ||
<div class="col-md-12"> | <div class="col-md-12"> | ||
Scroll down for more<br> | Scroll down for more<br> | ||
| − | <a href="#Project- | + | <a href="#Project-Description" class="btn btn-circle btn-transparent page-scroll"> |
<i class="fa fa-angle-double-down"></i> | <i class="fa fa-angle-double-down"></i> | ||
</a> | </a> | ||
| Line 275: | Line 275: | ||
| − | <div id="Project- | + | <div id="Project-Description"> |
<div class="container"> | <div class="container"> | ||
<div class="row col-md-12"> | <div class="row col-md-12"> | ||
<h1> | <h1> | ||
| − | Project | + | Project Description |
</h1> | </h1> | ||
<p style="text-align: center;"><span style="font-size:72px;"><span class="fa fa-headphones" style="color:rgb(0, 0, 0);"></span></span></p> | <p style="text-align: center;"><span style="font-size:72px;"><span class="fa fa-headphones" style="color:rgb(0, 0, 0);"></span></span></p> | ||
Revision as of 09:18, 2 October 2015
Project Description
Visit the Synthesizers' stand during Giant Jamboree! Get tuned in and listen to our podcast explaining our project!
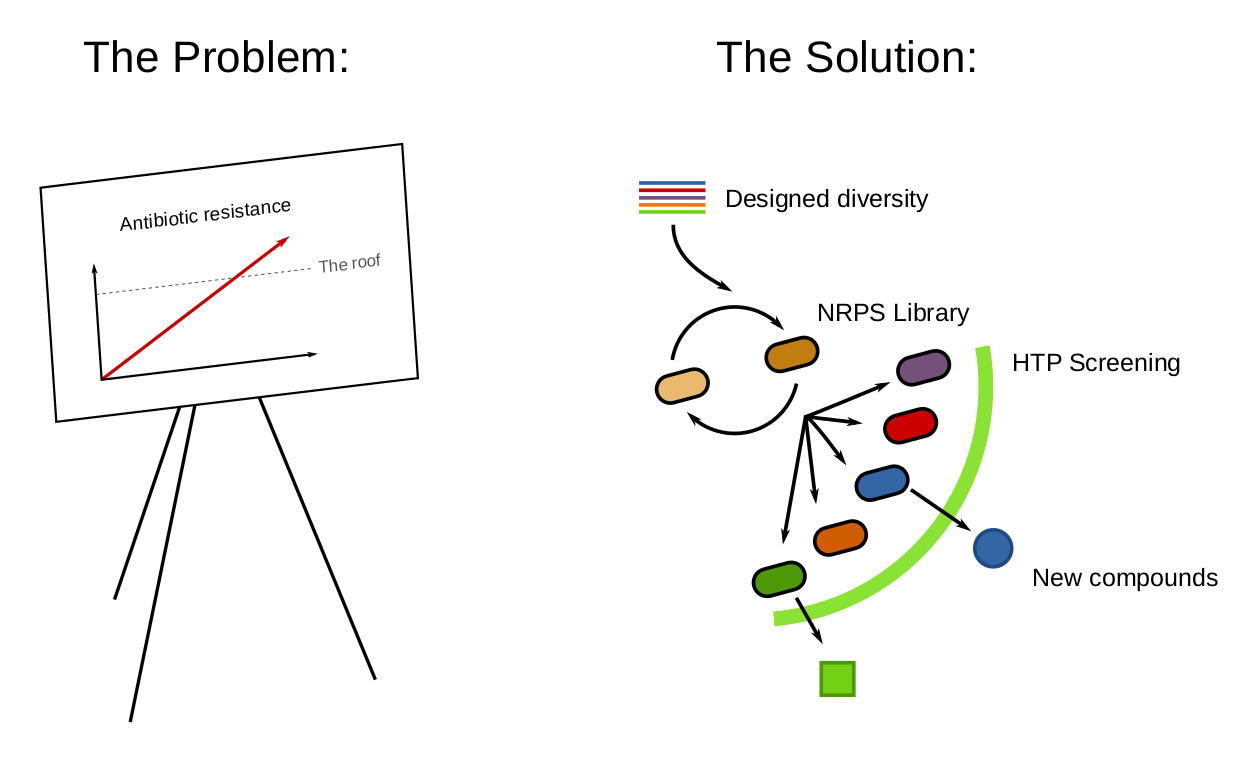
Figure 1
Non-ribosomal peptides have important anti-bacterial, anti-cancer, and immunosuppressive biological activities. They are synthesized by modular, high molecular weight enzymes that assemble more than 500 different amino acid substrates in an assembly line manner. For this reason, synthetic biologists have tried to engineer these proteins and to switch modules to create analogs and novel natural products, but with little success. Despite being modular, the interactions between modules have evolved to be highly specific, making synthetic Non-Ribosomal Peptide Synthases (NRPS) a challenge to engineer. Instead of switching modules we introduced a recombination system targeting oligo integration in Bacillus subtilis. We used the recombineering system to alter the active sites determining substrate specificity, thereby creating variants of antibiotics. Our focus was the tyrocidine antibiotic, which cannot be used intravenously due to its toxicity. Our goal is to create new analogs through multiplex automated genome engineering to reduce toxicity.
Department of Systems Biology
Søltofts Plads 221
2800 Kgs. Lyngby
Denmark
P: +45 45 25 25 25
M: dtu-igem-2015@googlegroups.com