Difference between revisions of "Team:Nagahama/Medal Parts"
(→Medal parts) |
(→銅) |
||
| Line 14: | Line 14: | ||
| − | others | + | others: |
| + | |||
[http://parts.igem.org/Part:BBa_K1653001] | [http://parts.igem.org/Part:BBa_K1653001] | ||
[http://parts.igem.org/Part:BBa_K1653002] | [http://parts.igem.org/Part:BBa_K1653002] | ||
| Line 46: | Line 47: | ||
[http://parts.igem.org/Part:BBa_K1653033] | [http://parts.igem.org/Part:BBa_K1653033] | ||
[http://parts.igem.org/Part:BBa_K1653034] | [http://parts.igem.org/Part:BBa_K1653034] | ||
| − | |||
| − | |||
==銀== | ==銀== | ||
Revision as of 05:59, 13 September 2015
Contents
Medal parts
銅
New Part
ispA [http://parts.igem.org/Part:BBa_K1653003 parts page]
ispA(from Escherichia coli JM109)
ispA encodes geranyl diphosphate synthase and farnesyl diphosphate synthase.Farnesyl diphosphate synthase can utilize both dimethylallyl and geranyl diphosphates as substrates, generating geranyl and farnesyl diphosphate, respectively. Therefore the enzyme can catalyze two sequential reactions in the polyisoprenoid biosynthetic pathway.
others:
[http://parts.igem.org/Part:BBa_K1653001] [http://parts.igem.org/Part:BBa_K1653002] [http://parts.igem.org/Part:BBa_K1653004] [http://parts.igem.org/Part:BBa_K1653005] [http://parts.igem.org/Part:BBa_K1653006] [http://parts.igem.org/Part:BBa_K1653007] [http://parts.igem.org/Part:BBa_K1653008] [http://parts.igem.org/Part:BBa_K1653009] [http://parts.igem.org/Part:BBa_K1653010] [http://parts.igem.org/Part:BBa_K1653011] [http://parts.igem.org/Part:BBa_K1653012] [http://parts.igem.org/Part:BBa_K1653013] [http://parts.igem.org/Part:BBa_K1653014] [http://parts.igem.org/Part:BBa_K1653015] [http://parts.igem.org/Part:BBa_K1653016] [http://parts.igem.org/Part:BBa_K1653017] [http://parts.igem.org/Part:BBa_K1653018] [http://parts.igem.org/Part:BBa_K1653019] [http://parts.igem.org/Part:BBa_K1653021] [http://parts.igem.org/Part:BBa_K1653022] [http://parts.igem.org/Part:BBa_K1653023] [http://parts.igem.org/Part:BBa_K1653024] [http://parts.igem.org/Part:BBa_K1653026] [http://parts.igem.org/Part:BBa_K1653027] [http://parts.igem.org/Part:BBa_K1653028] [http://parts.igem.org/Part:BBa_K1653029] [http://parts.igem.org/Part:BBa_K1653030] [http://parts.igem.org/Part:BBa_K1653031] [http://parts.igem.org/Part:BBa_K1653032] [http://parts.igem.org/Part:BBa_K1653033] [http://parts.igem.org/Part:BBa_K1653034]
銀
New Part2
ispA+MEP.dev[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1653025]
λPL+r.b.s.+ispA+MEP+×× (r.b.s.+dxs+r.b.s.+m-idi+r.b.s.+ispDF)
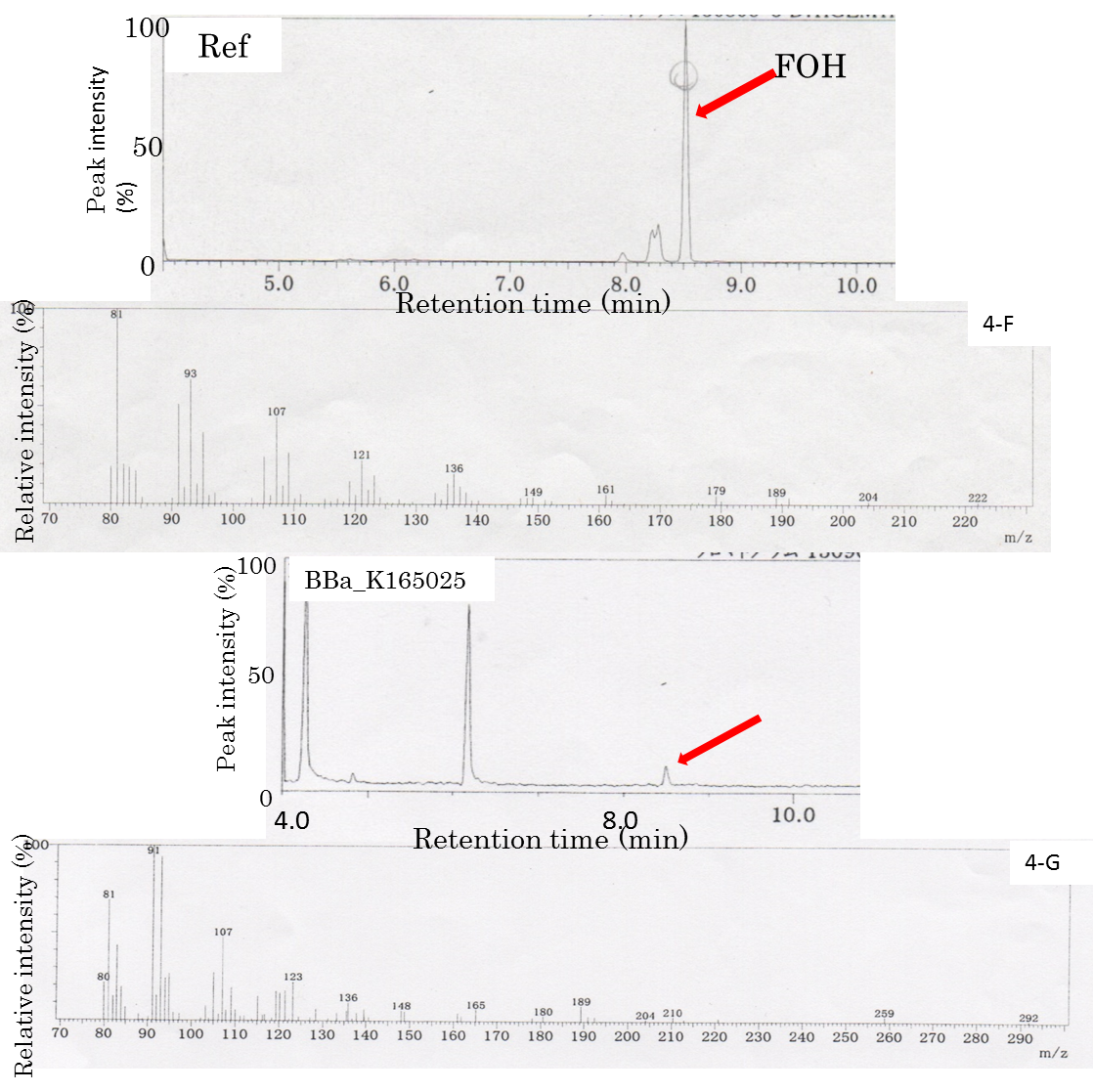
We submit new part(BBa_K165025) as producing FOH.
FOH is probably generated through FPP hydrolysis by endogenous phosphatases, which are induced by an increased intracellular FPP level Analogously, we hypothesized that E. coli could produce FOH under cellular conditions of an increased intracellular FPP level through metabolic engineering. A MEP pathway has been shown to synthesize IPP and DMAPP efficiently in E. coli. Because of its high hydrophobicity and low volatility, decane was chosen to extract and solubilize FOH from culture broth. The decane overlay in the two-phase culture did not affect growth, and FOH could be solubilized in the decane phase with negligible volatile loss. We adopt 1 mL of decane overlaid to 5 mL of culture broth. Two-phase culture of E. coli JM109 (BBa_K165025) was carried out in 2YT medium containing 1% glycerol at 29°C for 48 h. The decane phase of the two-phase culture was collected to analyze the FOH content by GC-MS. In the GC-MS analysis (Fig. 4A-G), there was a main peak at 8.5 min in the collected decane phase sample, which corresponded to the reference solution of the standard FOH compound dissolved in decane. Mass spectrometry confirmed that the peak at 8.5 min was FOH (Fig. 4-A). However, the peak was not observed in two-phase culture without introducing BBa_K165025. The formation of FOH from FPP was further confirmed by blocking FPP synthesis. In the GC-MS, the FOH peak was observed in E. coli JM109 (BBa_K165025) culture, whereas no peak was observed with transformed E. coli JM109. It was found that FOH need not only ispA(BBa_K165018) but also MEP(BBa_K165024) in E. coli. We submit new part(BBa_K165025) as producing FOH.
Gas Chromatography/Mass(GC/MS)
Fig4:The FOH standard solution (Ref) was used as a control. The peak corresponding to the FOH standard at 8.5 min is indicated by an arrow. The peak at 8.5 min was applied to GC/MS. The FOH standard solution (Ref) was used as a control. E. coli JM109(Bba_K165025) were compared with respect to FOH formation using GC-MS.
金
marA device
 [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1653020 Part's page]
[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1653020 Part's page]
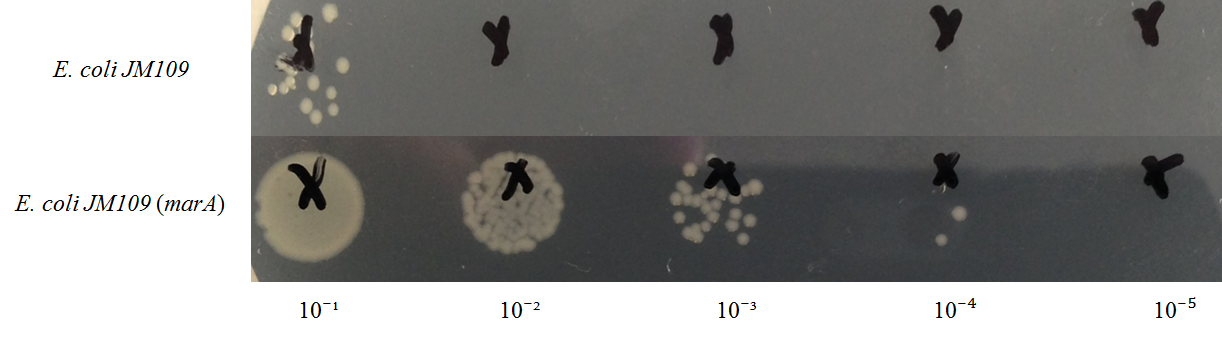
E. coli JM109 and E. coli JM109 (marA) were spotted on LBGMg agar plates in serial ten-fold dilutions (10‐¹~10⁻⁵), overlaid with geraniol solutions, and incubated at 30°C for 24 h. This figure shows that E. coli JM109 (marA) that overxpresses marA is more surviving on geraniol overlay plates than E. coli JM109 wild type.

A: E. coli JM109 (WT) + hexane B: E. coli JM109 (marA) +hexane C: E. coli JM109 (WT) + geraniol D: E. coli JM109 (marA) +geraniol
A and B increased almost the same on the plate. But C and D differed clealy. After 1 hour, C lost colony on the plate. In contrast, D could see colony all plates.
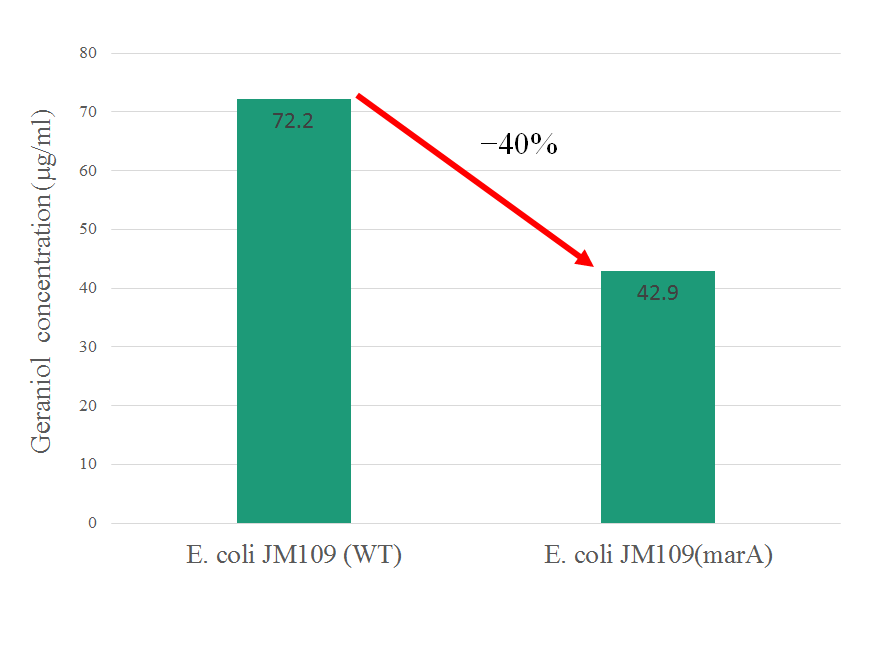
In our study, we confilmed that overexpressing of marA gives host E. coli high resistance against geraniol and reduce intracellular geraniol concentration.
(Gold Medal) In exsing part's information of marA, it gives E. coli resistance against kanamycin only. In this year, we confilmed that overepressing of marA gives E. coli resistance against geraniol as one of the terpene and decrease its intraceller concentration. This information is very beneficial for other iGEMers to production of organic substance that have toxicity using bacteria. So we improved the characterization of a previously existing BioBrick Part in our study. http://parts.igem.org/wiki/index.php?title=Part:BBa_K1230004 Part's number BBa_K1230004
NOTE
Note
In order to be considered for the Best Part Collection award, you must fill out this page.
Did your team make a lot of great parts? Is there a theme that ties all your parts together? Do you have more than 10 parts in this collection? Did you make a CRISPR collection, a MoClo collection, or a collection of awesome pigment parts? Describe your parts collection on this page, so the judges can evaluate you for the Best Part Collection award.