Difference between revisions of "Team:Bielefeld-CeBiTec/Results/HeavyMetals"
| Line 37: | Line 37: | ||
</figure> | </figure> | ||
</br> | </br> | ||
| − | + | </br></br>The tested heavy metal concentrations had no negative effect on <i>E. colis </i>growth. Moreover there is no significant difference between the curves with heavy metals and the controls. This first experiment showed us, in vivo characterization with these sensors under the tested heavy metal concentrations is possible. Most of our sensors were cultivated in the BioLector. Due to the accuracy of this device we could measure our sample in duplicates. </br> | |
| Line 50: | Line 50: | ||
<h1>Chromium</h1> | <h1>Chromium</h1> | ||
| − | + | </br></br> | |
| − | <h2><i>in vivo</i></h2> | + | <h2><i>in vivo</i></h2></br></br> |
| + | Our sensor for chromium detection consists of ChrB the repressor and the chromate specific promoter ChrP. The promoter is regulated by the ChrB, which binds Cr-ions. Behind the promoter is a sfGFP for detection of a fluorescence signal. | ||
| + | <i>In vivo</i> we could show that the addition of different concentrations of chromium have different effects to transcription of sfGFP. </br></br> | ||
<figure style="width: 600px"> | <figure style="width: 600px"> | ||
| − | <a href="https://static.igem.org/mediawiki/2015/8/82/Bielefeld-CeBiTec_Biolector_chromium.jpg" data-lightbox="heavymetals" data-title=" | + | <a href="https://static.igem.org/mediawiki/2015/8/82/Bielefeld-CeBiTec_Biolector_chromium.jpg" data-lightbox="heavymetals" data-title="Time course of the induction of a chromium biosensor with sfGFP for different chromium concentrations in vivo. The data are measured with BioLector and normalized on OD600. Error bars represent the standard deviation of two biological replicates. ."><img src="https://static.igem.org/mediawiki/2015/8/82/Bielefeld-CeBiTec_Biolector_chromium.jpg" alt="Adjusting the detection limit"></a> |
| − | <figcaption> | + | <figcaption>Time course of the induction of a chromium biosensor with sfGFP for different chromium concentrations in vivo. The data are measured with BioLector and normalized on OD600. Error bars represent the standard deviation of two biological replicates.</figcaption> |
</figure> | </figure> | ||
<figure style="width: 600px"> | <figure style="width: 600px"> | ||
| − | <a href="https://static.igem.org/mediawiki/2015/7/73/Bielefeld-CeBiTec_Biolector_chromium_Balkendiagramm.jpg" data-lightbox="heavymetals" data-title=" | + | <a href="https://static.igem.org/mediawiki/2015/7/73/Bielefeld-CeBiTec_Biolector_chromium_Balkendiagramm.jpg" data-lightbox="heavymetals" data-title="Fluorescence levels at three different stages of cultivation. Shown are levels after 60 minutes, 150 minutes and 650 minutes. Error bars represent the standard deviation of three biological replicates."><img src="https://static.igem.org/mediawiki/2015/7/73/Bielefeld-CeBiTec_Biolector_chromium_Balkendiagramm.jpg" alt="Adjusting the detection limit"></a> |
| − | <figcaption> | + | <figcaption>Fluorescence levels at three different stages of cultivation. Shown are levels after 60 minutes, 150 minutes and 650 minutes. Error bars represent the standard deviation of three biological replicates.</figcaption> |
</figure> | </figure> | ||
| + | </br>Our data lead to the conclusion that in a cell based system it is possible to detect chromium. | ||
| + | In contrast to our expectations with higher chromium concentrations we got lower fluorescence levels. These observations needed further investigation. </br> | ||
<h2><i>in vitro</i></h2> | <h2><i>in vitro</i></h2> | ||
<figure style="width: 600px"> | <figure style="width: 600px"> | ||
| − | <a href="https://static.igem.org/mediawiki/2015/9/99/Bielefeld-CeBiTec_Influence_of_chromium_on_the_cell_extract.jpeg" data-lightbox="heavymetals" data-title=" | + | <a href="https://static.igem.org/mediawiki/2015/9/99/Bielefeld-CeBiTec_Influence_of_chromium_on_the_cell_extract.jpeg" data-lightbox="heavymetals" data-title=" Influence of different chromium concentrations on our crude cell extract. Error bars represent the standard deviation of three biological replicates."><img src="https://static.igem.org/mediawiki/2015/9/99/Bielefeld-CeBiTec_Influence_of_chromium_on_the_cell_extract.jpeg" alt="Adjusting the detection limit"></a> |
| − | <figcaption> | + | <figcaption>Influence of different chromium concentrations on our crude cell extract. Error bars represent the standard deviation of three biological replicates.</figcaption> |
</figure> | </figure> | ||
<figure style="width: 600px"> | <figure style="width: 600px"> | ||
| − | <a href="https://static.igem.org/mediawiki/2015/d/d0/Bielefeld-CeBiTec_induction_chromium_in_chrB-cell-extract.jpeg" data-lightbox="heavymetals" data-title=" | + | <a href="https://static.igem.org/mediawiki/2015/d/d0/Bielefeld-CeBiTec_induction_chromium_in_chrB-cell-extract.jpeg" data-lightbox="heavymetals" data-title="Chromium specific cell extract made from <i>E. coli</i> cells which already expressed the repressor before cell extract production. Induction with different chromium concentrations. Error bars represent the standard deviation of three biological replicates."><img src="https://static.igem.org/mediawiki/2015/d/d0/Bielefeld-CeBiTec_induction_chromium_in_chrB-cell-extract.jpeg" alt="Adjusting the detection limit"></a> |
| − | <figcaption> | + | <figcaption>Chromium specific cell extract made from <i>E. coli</i> cells which already expressed the repressor before cell extract production. Induction with different chromium concentrations. Error bars represent the standard deviation of three biological replicates.</figcaption> |
</figure> | </figure> | ||
<figure style="width: 600px"> | <figure style="width: 600px"> | ||
| − | <a href="https://static.igem.org/mediawiki/2015/1/1e/Bielefeld-CeBiTec_correction_induction_chromium_in_chrB-cell-extract.jpeg" data-lightbox="heavymetals" data-title=" | + | <a href="https://static.igem.org/mediawiki/2015/1/1e/Bielefeld-CeBiTec_correction_induction_chromium_in_chrB-cell-extract.jpeg" data-lightbox="heavymetals" data-title="Chromium specific cell extract made from <i>E. coli</i> cells which already expressed the repressor before cell extract production. Induction with different chromium concentrations. Error bars represent the standard deviation of three biological replicates.Data are normalised on chromiums influence to the cell extrat."><img src="https://static.igem.org/mediawiki/2015/1/1e/Bielefeld-CeBiTec_correction_induction_chromium_in_chrB-cell-extract.jpeg" alt="Adjusting the detection limit"></a> |
| − | <figcaption> | + | <figcaption>Chromium specific cell extract made from <i>E. coli</i> cells which already expressed the repressor before cell extract production. Induction with different chromium concentrations. Error bars represent the standard deviation of three biological replicates.Data are normalised on chromiums influence to the cell extrat.</figcaption> |
</figure> | </figure> | ||
Revision as of 04:20, 14 September 2015
Heavy Metals
Zusammenfassung in ganz wenigen Worten.

Arsenic
in vivo

in vitro
Chromium
in vivo
Our sensor for chromium detection consists of ChrB the repressor and the chromate specific promoter ChrP. The promoter is regulated by the ChrB, which binds Cr-ions. Behind the promoter is a sfGFP for detection of a fluorescence signal. In vivo we could show that the addition of different concentrations of chromium have different effects to transcription of sfGFP.

in vitro





Copper
in vivo


in vitro






Lead
in vivo


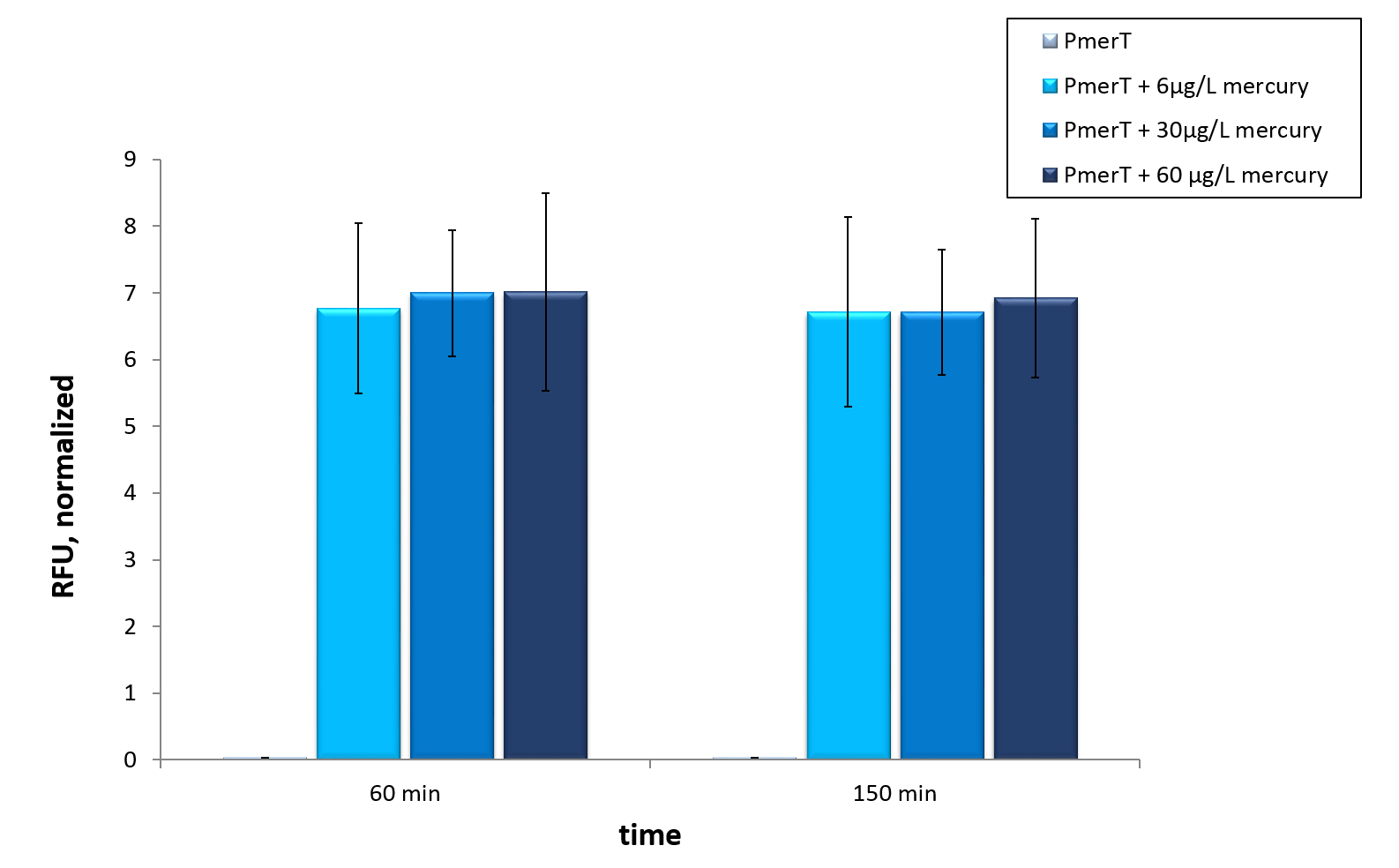
Mercury
in vivo


in vitro



Nickel
in vivo



