Difference between revisions of "Team:Nagahama/awarming"
(→Result and Discussion) |
(→Abstract) |
||
| Line 6: | Line 6: | ||
===Abstract=== | ===Abstract=== | ||
| + | Tokyo_Tech' plans at first was to integrate the chemotaxis experiment into our overall project. | ||
| + | Unfortunately, Tokyo_Tech were unable to obtain positive results from the gene that they constructed, before the Giant Jamboree. | ||
| + | Chemotaxis experiment is very difficult. | ||
| + | We helped Tokyo_Tech by the chemotaxis experiment. | ||
| + | We helped team Tokyo_Tech by advising these 2 about chemotaxis experiments. | ||
| + | 1. In order to increase the chemotactic activity of E. coli, the best concentration of agar is 0.3 %. | ||
| + | 2. It is better to directly stick the chip inside the agar and then inject the E .coli, than to place the E. coli on top of the agar | ||
| + | Also, we gave Tokyo_Tech the raw data along with the protocol of the experiments for assaying the chemotactic activity of a wild type E.coli. | ||
===Method=== | ===Method=== | ||
Latest revision as of 10:44, 18 September 2015
Contents
Swarming Support
We helped Tokyo_Tech.
Abstract
Tokyo_Tech' plans at first was to integrate the chemotaxis experiment into our overall project. Unfortunately, Tokyo_Tech were unable to obtain positive results from the gene that they constructed, before the Giant Jamboree. Chemotaxis experiment is very difficult. We helped Tokyo_Tech by the chemotaxis experiment. We helped team Tokyo_Tech by advising these 2 about chemotaxis experiments. 1. In order to increase the chemotactic activity of E. coli, the best concentration of agar is 0.3 %. 2. It is better to directly stick the chip inside the agar and then inject the E .coli, than to place the E. coli on top of the agar Also, we gave Tokyo_Tech the raw data along with the protocol of the experiments for assaying the chemotactic activity of a wild type E.coli.
Method
5 × M9 salts stock solution
6.4 % (w/v) Na2HPO4 × 7H2O
1.5 % (w/v) KH2PO4
0.25 % (w/v) NaCl
0.5 % (w/v) NH4Cl
M9 swarming agar
1.25 % (v/v) glycerol
20 % (v/v) 5 × M9 salts stock solution
0.1 % (v/v) of CaCl2 × 2H2O stock solution (20 mg/mL)
0.1 % (v/v) MgSO4 stock solution (0.12 g/mL)
0.03 % (w/v) thiamine
0.3 % (w/v) agar
tripton broth
1.0 % (w/v) tripton
1.0 % (w/v) NaCl
Swarming Assay
We assayed chemotaxis of E. coli against cadmium and aspartic acid on soft agar, containing M9 synthetic medium, on that plate E. coli can swim. Protocol 1.E. coli JM109 was cultured at 30℃ for 12 hours with shaking (50 rpm). 2.Aliquot of the culture was spotted on the center of agar plate. 3.10 mM L-aspartic acid (40 μl) or 100 mM cadmium chloride (4 μl) was spotted on 25 mm distant from the center of the agar plate. 4.The plate was standed for 5 min at RT. 5.The plate was incubated at 30 ℃.
Result and Discussion
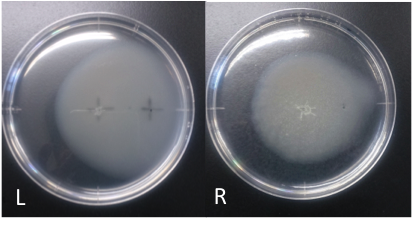
fig1:E. coli swarming toward aspartic acid L: aspartic acid (10 mM, 40 μl) R: ddH2O (40 μl) Incubation: 108 hours temperature: 30 ℃ E. coli cultured was spotted on the center of agar plate, and chemotaxis was spotted on 25 mm distant from the center. The swarming circle of aspartic acid (L) was larger than that of ddH2O (R), and the swarming area around aspartate acid (L) was more larger than that of ddH2O (R), suggesting that E. coli swarming to aspartate acid. This experiment indicate that E. coli might have positive chemotaxis toward aspartic acid.
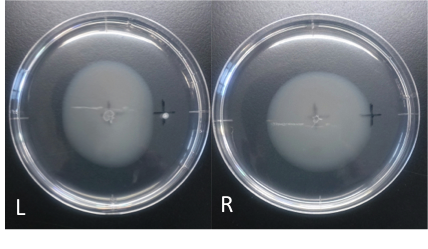
fig2:E. coli hate cadmium. L: cadmium(100 mM, 4 μl) R: ddH2O (40 μl) Incubation: 110 hours temperature: 30 ℃ E. coli cultured was spotted on the center of agar plate, and chemotaxis was spotted on 25 mm distant from the center. The swarming circle of cadmium (L) was smaller than that of ddH2O (R), and the swarming area around cadmium (L) was more smaller than that of ddH2O (R), suggesting that E. coli hate cadmium. This experiment indicate that E. coli might have negative chemotaxis against cadmium.