Difference between revisions of "Team:Bielefeld-CeBiTec/Project/PRIA"
| Line 47: | Line 47: | ||
<div class="col-md-6"> | <div class="col-md-6"> | ||
<h3>Immobilized Repressor</h3> | <h3>Immobilized Repressor</h3> | ||
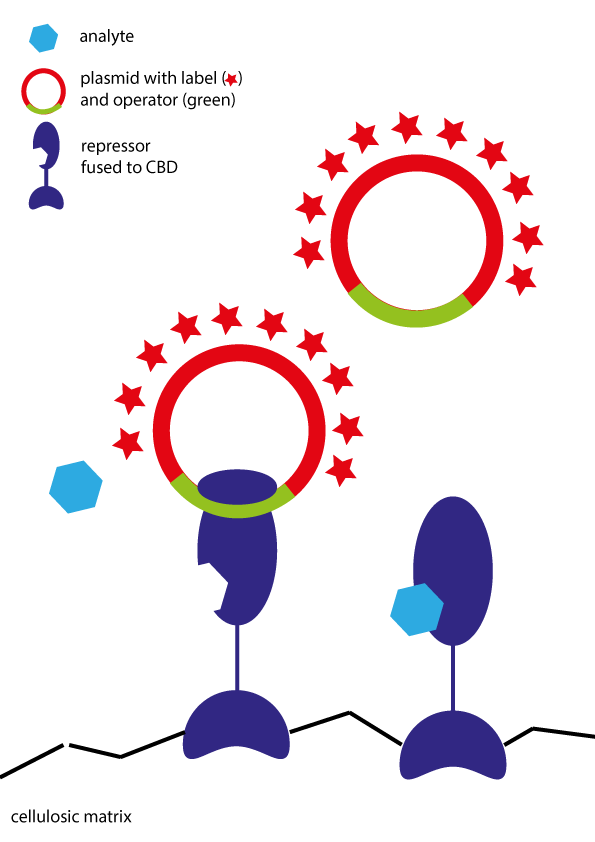
| − | <p>The basis of our approach is to build on fusion proteins of the repressor proteins with cellulose-binding domains (CBD). By adding these proteins to paper, the repressor is immobilized. DNA which is fluorophore-labeled (in our case: Cy3-labeled) is added to the proteins. The repressors bind to the DNA on the operator site. After analyte addition, DNA is released and the loss of signal can be detected (see <a href="https://static.igem.org/mediawiki/2015/b/b9/PRIA_immobProtein_imagesonly.gif">image</a>). </p | + | <p>The basis of our approach is to build on fusion proteins of the repressor proteins with cellulose-binding domains (CBD). By adding these proteins to paper, the repressor is immobilized. DNA which is fluorophore-labeled (in our case: Cy3-labeled) is added to the proteins. The repressors bind to the DNA on the operator site. After analyte addition, DNA is released and the loss of signal can be detected (see <a href="https://static.igem.org/mediawiki/2015/b/b9/PRIA_immobProtein_imagesonly.gif">image</a>). </p></br></br> |
<figure><a href="https://static.igem.org/mediawiki/2015/b/b9/PRIA_immobProtein_imagesonly.gif" data-lightbox="projectPRIA" data-title="Illustration of PRIA with repressor protein bound to a cellulose matrix. Due to the addition of the analyte, the repressor is released. The signal measured is generated by the release of plasmid DNA." > | <figure><a href="https://static.igem.org/mediawiki/2015/b/b9/PRIA_immobProtein_imagesonly.gif" data-lightbox="projectPRIA" data-title="Illustration of PRIA with repressor protein bound to a cellulose matrix. Due to the addition of the analyte, the repressor is released. The signal measured is generated by the release of plasmid DNA." > | ||
<img src="https://static.igem.org/mediawiki/2015/b/b9/PRIA_immobProtein_imagesonly.gif" alt="Sorry, cannot load this file at the moment" width="350 px"></a> | <img src="https://static.igem.org/mediawiki/2015/b/b9/PRIA_immobProtein_imagesonly.gif" alt="Sorry, cannot load this file at the moment" width="350 px"></a> | ||
Revision as of 00:50, 19 September 2015
Plasmid Repressor Interaction Assay
A Cell-free Detection System based on two purified Components
Introduction
Cell-free protein synthesis is a promising approach for our biosensors. However, due to the need for transcription and translation it takes up to one hour until a signal can be detected. Furthermore, the complex machinery could be prone to disturbances (e.g. high pH, glycerol). Therefore, we decided to test alternative designs. By avoiding protein synthesis and working with purified proteins and DNA, we developed a simplified biosensor that is very fast. The short time span is of significant advantage for biosensors in the open field, e.g. in testing beverages for date rape drugs.
In our developed assay, a repressor protein forms a complex with a plasmid containing the corresponding operator sequence. The repressor changes its conformation upon binding of the targeted substrate (further referred to as "analyte") and the bond to the DNA is broken. This disruption will be detected via a loss of a fluorescence signal caused by elution of labeled protein or DNA. We call this system Plasmid Repressor Interaction Assay (PRIA). In comparison to the cell extract, this method does require neither transcription nor translation of a reporter protein. Therefore, the signal can be measured faster than in cell extracts and cellular systems. Another disadvantage of cell-based biosensors is that microorganisms are more sensible against toxic substances. This problem can be solved using this assay. A cell-free system can tolerate a higher amount of these substances. According to Siddiki et al. who work with a similar system based on immobilized DNA and a GFP-fused repressor measurement of arsenic and cadmium contamination of water, it was possible to get a signal within 15 to 30 minutes (Siddiki et al. 2011). Thereby this approach meets one of the requirements often mentioned by potential users in our survey on synthetic biology and biosensors: speed.
Furthermore, PRIA works with just two purified components. So there is no risk of releasing genetically modified organisms (GMOs) into the environment as we do not work with them. Our system is not even subject to the regulations of the German "Gentechnikgesetz" (Genetic engineering Act). This system should be robust against environmental factors, as long as they do not denaturize the protein and show increased stability and durability compared to conventional biosensors, since there are no metabolic pathways involved. Dr. Mathias Keller, Supervisor of laboratories regarding biosafety issues in the district East Westphalia Lippe, verifies our hypothesis (Dr. Mathias Keller from the district government, authorized for judging biosafety of the laboratories in the district East Westphalia Lippe).
Aim
In this part of our project, we developed a sensor based on the disruption of a repressor-plasmid binding. Paper-based systems are increasing in popularity because they are cheap and easily renewable. Therefore, we wanted to establish a paper-based test strip, which enables swift detection of analytes (e.g. copper, arsenic) by a loss of fluorescence signal.
Strategies
For our project we wanted to implement a system based on repressor protein-operator DNA interaction described on the project design's page. By adding the analyte solution, the interaction should be dissolved. We started our experiments with a good characterized model system for repression and derepression: the lac operon and its repressor LacI (Santillan et al.). LacI immobilized on a Ni-NTA agarose column delivered the first proof of concept: Plasmid DNA containing the lac promoter was bound to LacI and could be detected in the elution fraction upon addition of Isopropyl-β-D-thiogalactopyranosid (IPTG) to the column. The applied protocol was similar to that of protein-protein interaction studies. We wanted to transfer this system from an impractical Ni-NTA agarose column to paper as paper-based systems are easy to use and cheap. There are two possible options to do so. The first one would be to immobilize the repressor protein on paper, the second is to immobilize the DNA with the operator on paper.
Immobilized Repressor
The basis of our approach is to build on fusion proteins of the repressor proteins with cellulose-binding domains (CBD). By adding these proteins to paper, the repressor is immobilized. DNA which is fluorophore-labeled (in our case: Cy3-labeled) is added to the proteins. The repressors bind to the DNA on the operator site. After analyte addition, DNA is released and the loss of signal can be detected (see image).
Immobilized DNA
The immobilization of ssDNA on paper has been reported before (Araújo et al. 2012). We adapted this method to enable the immobilization of dsDNA and combined it with an approach based on the measurement of the disruption of the binding of a GFP-tagged repressor protein to immobilized DNA as reported before (Siddiki et al. 2011). In this strategy, we add GFP-fused repressor protein which binds to the immobilized DNA. After analyte addition the interaction between operator DNA and GFP-fused DNA would be dissolved and the signal of GFP-fused protein is reduced after the elution step.

Summary
In summary, cell-free systems are universally usable and less sensible to toxic substances. In comparison to cellular systems, the signals are faster to detect. Moreover, they fulfill the need of biosafety. Therefore, we wanted to create a system based on repressor protein-operator DNA interaction. First, we established the proof of concept for PRIA on Ni-NTA agarose with our model system lacO-LacI. Then we wanted to show that the immobilization of dsDNA on paper is feasible. Paper-based systems become more popular due to their easiness to use and the low cost. We pursued two strategies within the establishment of our PRIA assay: one is based on GFP-fused repressor protein and immobilized DNA on paper, the other one is based on fluorophore-tagged DNA and immobilized repressor protein tagged to a cellulose-binding domain. In the results we implemented this assay on Ni-NTA and brought the aspects of the assay and paper together. We optimized a protocol that allows the establishment of the protein DNA complex on paper.
References
Araújo, Ana Caterina; Song, Yajing; Lundeberg, Joakim; Stahl, Patrik L.; Brumer, Harry (2012): Activated Paper Surfaces for the Rapid Hybridization of DNA through Capillary Transport. In Anal. Chem. 2012, 84, pp. 3311-3317. DOI: 10.1021/ac300025v
Lehtiö, Janne; Wernerús, Henrik; Samuelson, Patrik; Teeri, Tuula T.; Stahl, Stefan (2001): Directed immobillization of recombinant staphylococci on cotton fibers by functional display of a fungal cellulose-binding domain. In FEMS Microbiol Lett. 2001, 195(2), pp. 197-204. DOI: 10.1111/j.1574-6968.2001.tb10521.x 197-204
Santillan, M.; Mackey, M. C. (2008): Quantitative approaches to the study of bistability in the lac operon of Escherichia coli. In Journal of The Royal Society Interface 5 (Suppl_1), pp. S29-S39. DOI: 10.1098/rsif.2008.0086.focus.
Siddiki, Mohammad Shohel Rana; Kawakami, Yasunari; Ueda, Shunsaku; Maeda, Isamu (2011): Solid Phase Biosensors for Arsenic or Cadmium Composed of A trans Factor and cis Element Complex.In Sensors 2011, 11, pp. 10063-10073. 10.3390/s111110063
German Genetic Engineering Act,http://www.gesetze-im-internet.de/bundesrecht/gentg/gesamt.pdf, last checked: 9/18/2015



