Difference between revisions of "Team:SJTU-BioX-Shanghai/Composite Part"
| Line 4: | Line 4: | ||
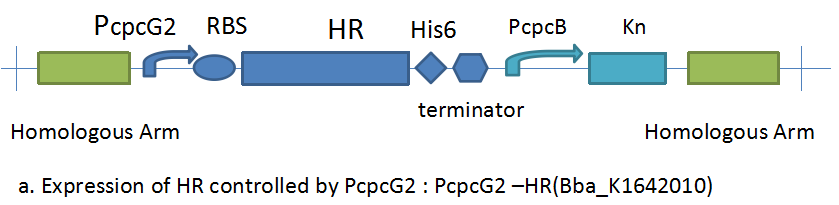
==PcpcG2-HR[[http://parts.igem.org/Part:BBa_K1642010 BBa_K1642010]]== | ==PcpcG2-HR[[http://parts.igem.org/Part:BBa_K1642010 BBa_K1642010]]== | ||
| − | [[File:SJTUB PcpcG2-HR.png | 500px | middle| frameless ]] | + | |
| + | <center>{{ Template:SJTU-BioX-Shanghai/Figure | ||
| + | | figure = [[File:SJTUB PcpcG2-HR.png | 500px | middle| frameless ]] | ||
| + | | float = middle | ||
| + | | id = 2.2.1 | ||
| + | | label = Identification by Enzyme Digestion | ||
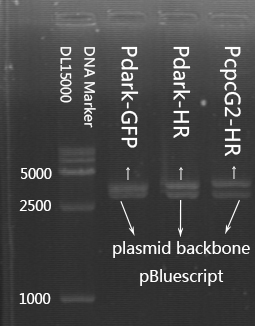
| + | | descr = The backbone of plasmid pBluescript is about 3000bp. The fragments of Pdark-HR, Pdark-HR, PcpcG2-HR are about 3370bp, 3570bp, 3770bp. | ||
| + | }}</center> | ||
===Process controlled by PcpcG2-HR=== | ===Process controlled by PcpcG2-HR=== | ||
| Line 12: | Line 19: | ||
| figure = [[File:SJTUB biode(2).png| 410px | frameless ]] | | figure = [[File:SJTUB biode(2).png| 410px | frameless ]] | ||
| float = middle | | float = middle | ||
| − | | id = | + | | id = 2.2.2 |
| label = Biodesalination process controlled by PcpcG2. | | label = Biodesalination process controlled by PcpcG2. | ||
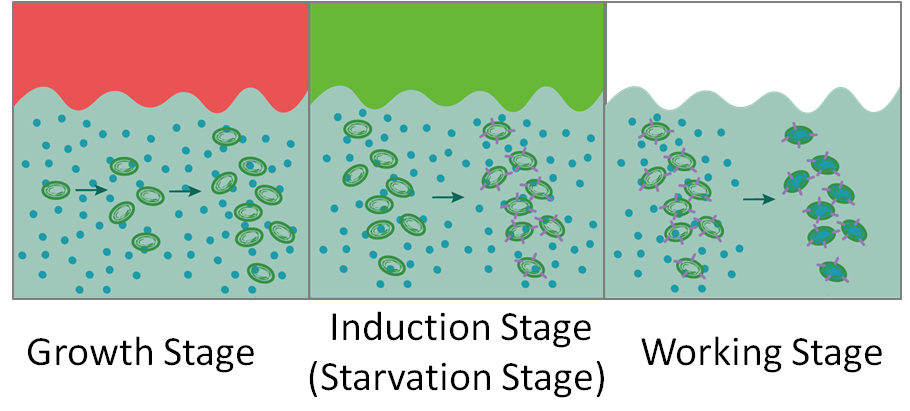
| descr = In the growth stage, cyanobacteria absorb energy from red light and grow to a certain density; In the induction stage, green-light induces the expression of halorhodopsin, and additionally pushes cyano bacteria to starvation status; In the working stage, engineered cyanobacteria absorb sodium chloride under natural light and cyanobacteria regain energy. | | descr = In the growth stage, cyanobacteria absorb energy from red light and grow to a certain density; In the induction stage, green-light induces the expression of halorhodopsin, and additionally pushes cyano bacteria to starvation status; In the working stage, engineered cyanobacteria absorb sodium chloride under natural light and cyanobacteria regain energy. | ||
| Line 21: | Line 28: | ||
| figure = [[File:SJTUB 0917.jpg| 410px | frameless ]] | | figure = [[File:SJTUB 0917.jpg| 410px | frameless ]] | ||
| float = middle | | float = middle | ||
| − | | id = | + | | id = 2.2.3 |
| label = Identification by Enzyme Digestion | | label = Identification by Enzyme Digestion | ||
| descr = The backbone of plasmid pBluescript is about 3000bp. The fragments of Pdark-HR, Pdark-HR, PcpcG2-HR are about 3370bp, 3570bp, 3770bp. | | descr = The backbone of plasmid pBluescript is about 3000bp. The fragments of Pdark-HR, Pdark-HR, PcpcG2-HR are about 3370bp, 3570bp, 3770bp. | ||
| Line 28: | Line 35: | ||
===Transfomation=== | ===Transfomation=== | ||
| − | PcpcG2 controls the expression of halorhodopsin in ''Synnechosystis sp.'' strain PCC 6803 through composite part PcpcG2-HR (BBa_K1642010). We applied kanamycin to screen the transformants and test the success of transformation by colony PCR. The results of colony PCR are shown in Figure | + | PcpcG2 controls the expression of halorhodopsin in ''Synnechosystis sp.'' strain PCC 6803 through composite part PcpcG2-HR (BBa_K1642010). We applied kanamycin to screen the transformants and test the success of transformation by colony PCR. The results of colony PCR are shown in Figure 4. |
<center>{{ Template:SJTU-BioX-Shanghai/Figure | <center>{{ Template:SJTU-BioX-Shanghai/Figure | ||
| figure = [[File:SJTUB cp PcpcG2-HR.jpg | 300px]] | | figure = [[File:SJTUB cp PcpcG2-HR.jpg | 300px]] | ||
| − | | id = | + | | id = 2.2.4 |
| label = Colony PCR of PcpcG2-HR. | | label = Colony PCR of PcpcG2-HR. | ||
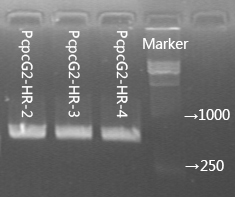
| descr = The forward primer is located in the upstream homologous arm and the reverse one is located in HR. The target fragments for colony PCR of PcpcG2-HR are approximately 370bp and 570bp respectively. The number at the tail of the sample represents our number for the colonies of transformant. | | descr = The forward primer is located in the upstream homologous arm and the reverse one is located in HR. The target fragments for colony PCR of PcpcG2-HR are approximately 370bp and 570bp respectively. The number at the tail of the sample represents our number for the colonies of transformant. | ||
| Line 42: | Line 49: | ||
| figure = [[File:SJTUB biode(41).jpg| 410px | frameless ]] | | figure = [[File:SJTUB biode(41).jpg| 410px | frameless ]] | ||
| float = middle | | float = middle | ||
| − | | id = | + | | id = 2.2.5 |
| label = Biodesalination assay of the process controlled by PcpcG2. | | label = Biodesalination assay of the process controlled by PcpcG2. | ||
| descr = The start of induction stage is at -12h and the times of taking samples are at -12h, 4h, 12h and 24h. | | descr = The start of induction stage is at -12h and the times of taking samples are at -12h, 4h, 12h and 24h. | ||
| Line 56: | Line 63: | ||
| figure = [[File:SJTUB biode(3).png| 410px | frameless ]] | | figure = [[File:SJTUB biode(3).png| 410px | frameless ]] | ||
| float = middle | | float = middle | ||
| − | | id = | + | | id = 2.2.6 |
| label = Biodesalination process controlled by Pdark. | | label = Biodesalination process controlled by Pdark. | ||
| descr = In the growth Stage, cyanobacteria grow to a log-phase; in the induction stage, darkness induces the expression of halorhodopsin, and additionally pushes cyanobacteria to starvation status; in the working stage, engineered cyanobacteria absorb sodium chloride under natural light. | | descr = In the growth Stage, cyanobacteria grow to a log-phase; in the induction stage, darkness induces the expression of halorhodopsin, and additionally pushes cyanobacteria to starvation status; in the working stage, engineered cyanobacteria absorb sodium chloride under natural light. | ||
| Line 70: | Line 77: | ||
<center>{{ Template:SJTU-BioX-Shanghai/Figure | <center>{{ Template:SJTU-BioX-Shanghai/Figure | ||
| figure = [[File:SJTUB cp Pdark-HR.jpg | 300px]] | | figure = [[File:SJTUB cp Pdark-HR.jpg | 300px]] | ||
| − | | id = | + | | id = 2.2.7 |
| label = Colony PCR of Pdark-HR. | | label = Colony PCR of Pdark-HR. | ||
| descr = The forward primer is located in the upstream homologous arm and the reverse one is located in HR. The target fragments for colony PCR of PcpcG2-HR are approximately 370bp and 570bp respectively. The number at the tail of the sample represents our number for the colonies of transformant. | | descr = The forward primer is located in the upstream homologous arm and the reverse one is located in HR. The target fragments for colony PCR of PcpcG2-HR are approximately 370bp and 570bp respectively. The number at the tail of the sample represents our number for the colonies of transformant. | ||
| Line 81: | Line 88: | ||
| figure = [[File:SJTUB_biode(51).jpg| 410px | frameless ]] | | figure = [[File:SJTUB_biode(51).jpg| 410px | frameless ]] | ||
| float = middle | | float = middle | ||
| − | | id = | + | | id = 2.2.8 |
| label = Biodesalination assay of the process controlled by Pdark. | | label = Biodesalination assay of the process controlled by Pdark. | ||
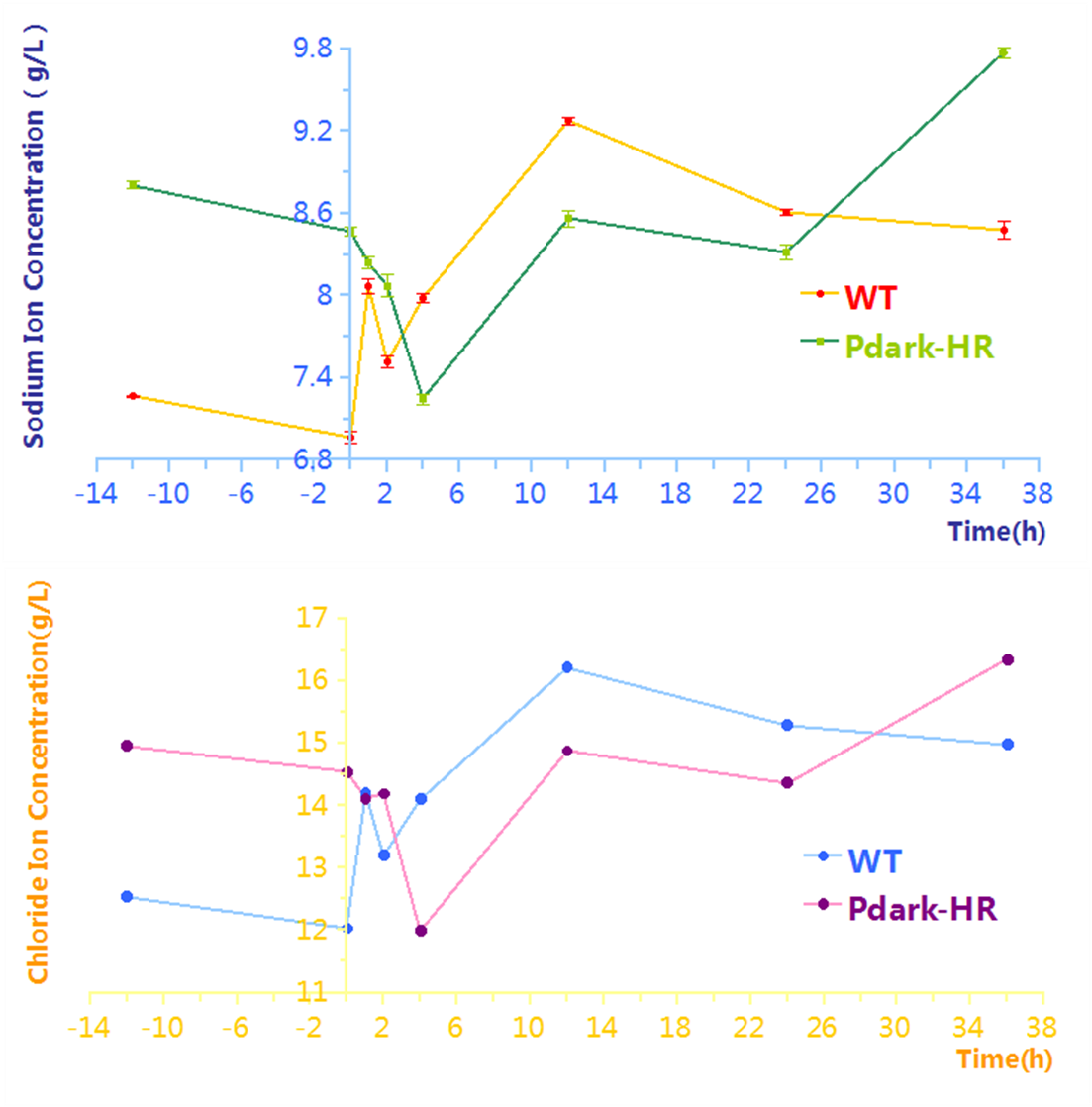
| descr = The start of induction stage is at -12h and the times of taking samples are at -12h, 0h, 1h, 2h, 4h, 12h, 24h and 36h. | | descr = The start of induction stage is at -12h and the times of taking samples are at -12h, 0h, 1h, 2h, 4h, 12h, 24h and 36h. | ||
| Line 90: | Line 97: | ||
| figure = [[File:SJTUB_biode(61).jpg| 410px | frameless ]] | | figure = [[File:SJTUB_biode(61).jpg| 410px | frameless ]] | ||
| float = middle | | float = middle | ||
| − | | id = | + | | id = 2.2.9 |
| label = Biodesalination assay of the process controlled by Pdark with more frequent sampling times. | | label = Biodesalination assay of the process controlled by Pdark with more frequent sampling times. | ||
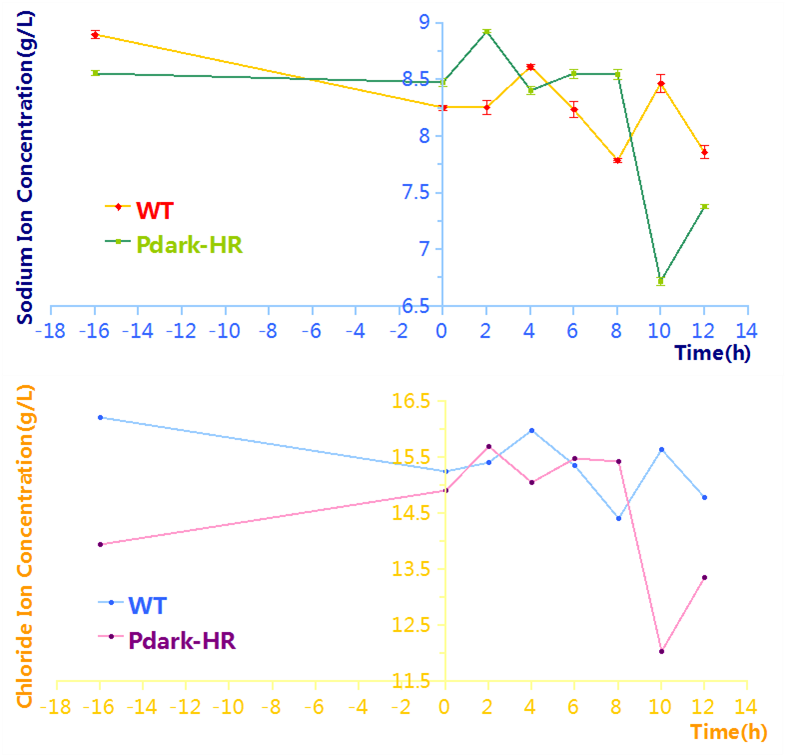
| descr = The start of induction stage is at -16h and the times of taking samples are at -16h, 0h, 2h, 2h, 4h, 6h, 8h,10h,12h. | | descr = The start of induction stage is at -16h and the times of taking samples are at -16h, 0h, 2h, 2h, 4h, 6h, 8h,10h,12h. | ||
Revision as of 01:19, 19 September 2015
Contents
PcpcG2-HRhttp://parts.igem.org/Part:BBa_K1642010 BBa_K1642010
Process controlled by PcpcG2-HR
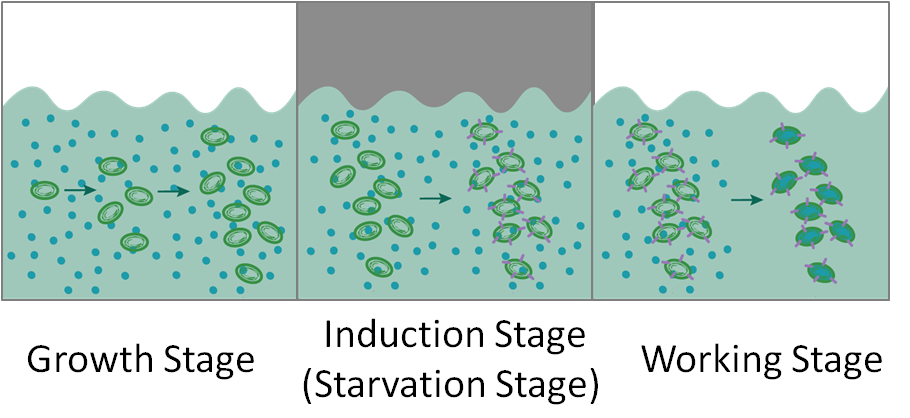
Based on PcpcG2 and halorhodopsin, we established a biodesalination system which relies on red and green light. The light colors in the three stages are red light, green light and white light respectively.
Figure 2.2.2 Biodesalination process controlled by PcpcG2.
In the growth stage, cyanobacteria absorb energy from red light and grow to a certain density; In the induction stage, green-light induces the expression of halorhodopsin, and additionally pushes cyano bacteria to starvation status; In the working stage, engineered cyanobacteria absorb sodium chloride under natural light and cyanobacteria regain energy.
Identification by Enzyme Digestion
The backbone of plasmid pBluescript is about 3000bp. The fragment of PcpcG2-HR is about 3770bp.
Transfomation
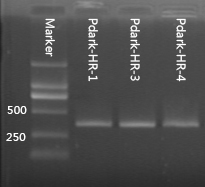
PcpcG2 controls the expression of halorhodopsin in Synnechosystis sp. strain PCC 6803 through composite part PcpcG2-HR (BBa_K1642010). We applied kanamycin to screen the transformants and test the success of transformation by colony PCR. The results of colony PCR are shown in Figure 4.
Figure 2.2.4 Colony PCR of PcpcG2-HR.
The forward primer is located in the upstream homologous arm and the reverse one is located in HR. The target fragments for colony PCR of PcpcG2-HR are approximately 370bp and 570bp respectively. The number at the tail of the sample represents our number for the colonies of transformant.
Assay on PcpcG2-HR
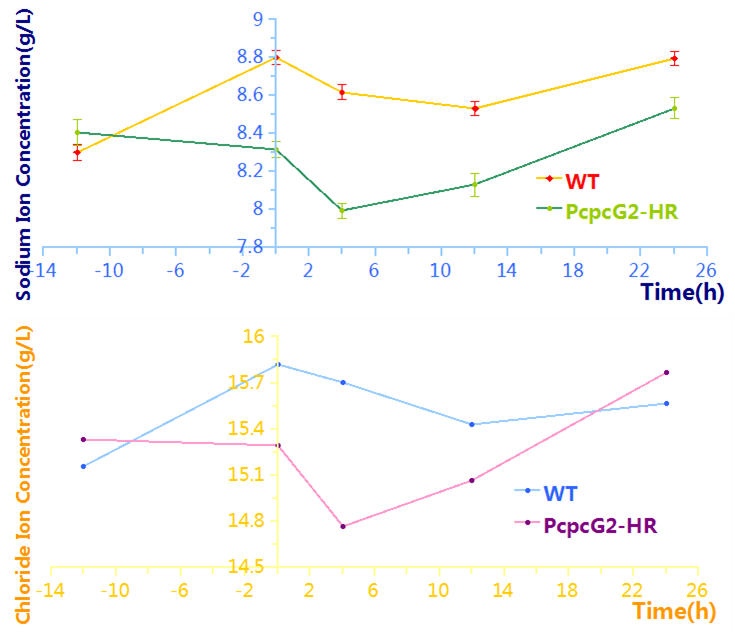
The effectiveness of this biodesalinaion process controlled by PcpcG2-HR is proved by determination of the concentrations of extracelluar sodium and chloride or desalination assay, which are shown in Figure 3. During the early time of working stage, there is an obvious decrease of concentration compared to that of Wild-type, which indicates that our biobrick (PcpcG2-HR, BBa_K1642010) really works in cyanobacteria under this biodesalination process! The following rise of concentration can be explained by the regain of energy under natural light in the working stage. Considering that we focus on the biodesalination process controlled by Pdark, we didn’t optimize this process.
Park-HRhttp://parts.igem.org/Part:BBa_K1642011 BBa_K1642011
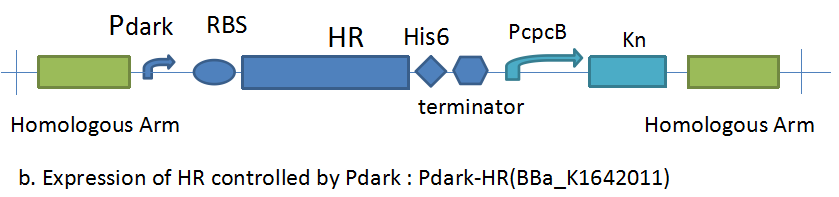
Process controlled by Pdark-HR
Based on Pdark and HR, we constructed an improved biodesalination system which depends only on the switch between white light and darkness. In the growth stage and working stage, we provide white light, while in the induced expression stage the light source is removed. Darkness leads to starvation and the starvation can inhibit active export of sodium, which is essential for biodesalination in the working stage.(described in section transport module). The biodesalination process controlled by Pdark is shown in Figure 3.
Figure 2.2.6 Biodesalination process controlled by Pdark.
In the growth Stage, cyanobacteria grow to a log-phase; in the induction stage, darkness induces the expression of halorhodopsin, and additionally pushes cyanobacteria to starvation status; in the working stage, engineered cyanobacteria absorb sodium chloride under natural light.
Identification by Enzyme Digestion
The result of identification by enzyme digestion is shown in Figure2.
The backbone of plasmid pBluescript is about 3000bp. The fragment of Pdark-HR is about 3570bp.
Transfomation
Pdark controls the expression of halorhodopsin in Synnechosystis sp. strain PCC 6803 through composite part Pdark-HR (BBa_K1642011). We applied kanamycin to screen the transformants and test the success of transformation by colony PCR. The results of colony PCR are shown in Figure 2.
Figure 2.2.7 Colony PCR of Pdark-HR.
The forward primer is located in the upstream homologous arm and the reverse one is located in HR. The target fragments for colony PCR of PcpcG2-HR are approximately 370bp and 570bp respectively. The number at the tail of the sample represents our number for the colonies of transformant.
Assay on Pdark-HR
The effectiveness of this biodesalination process controlled by Pdark-HR is proved by determination of the concentrations of extracellular sodium and chloride or desalination assay, which are shown in Figure 4. During the early time of working stage, there is an obvious decrease of concentration compared to that of wild-type, which proves the function of our biobrick(Park-HR, BBa_K1642011). An obvious decrease during the early time and a following rise are consistent with that of the process controlled by PcpcG2.
To figure out the limitation of desalination, we prolonged the length of the induction stage and adjusted the times of taking samples. The results are shown in Figure 5. The 6h in the working stage is approximately the minimum point.
The acquisition of the minimum point makes it possible to design a longer biodesalination process. We can extend this process by alternating induction stage (starvation stage) and working stage, make the cyanobacteria to experience starvation and regain of energy for more cycles, thus achieving more reduction of salinity. Moreover, if the length of the induction stage and working stage are approximately 12h, after growth stage this improved biodesalination system can be controlled by the natural alternation between day and night without any human intervention.