Team:DTU-Denmark/Project/Intein
Purpose
We did not succeed in expression of tyrocidine from Brevibacillus parabrevis in Bacillus subtilis [here]. This inspired us to think of alternative approaches for production of nonribosomal peptide drugs than through NRPS engineering. We came up with a peculiar idea of generating short cyclized peptides with similar length to tyrocidine by using self-splicing proteins. Inteins are such short self-splicing proteins that have no function in the proteins they are a part of, besides catalyzing their own excision after translation. The splicing makes a peptide bond between the two adjacent amino acids next to the inteins (Figure 1). If the N-intein and C-intein are placed at each end of the same polypeptide; cyclisation would give a cyclized protein - or in our case a short cyclic peptide cyclized from head-to-tail.
Achievements
- Designed constructs for expression of two short cyclic peptides using BioBrick BBa_K1362000 and two short oligo primers.
- Generated the cyclisation construct
- Analyzed constructs for cyclic peptides, but no products could be detected.
Background
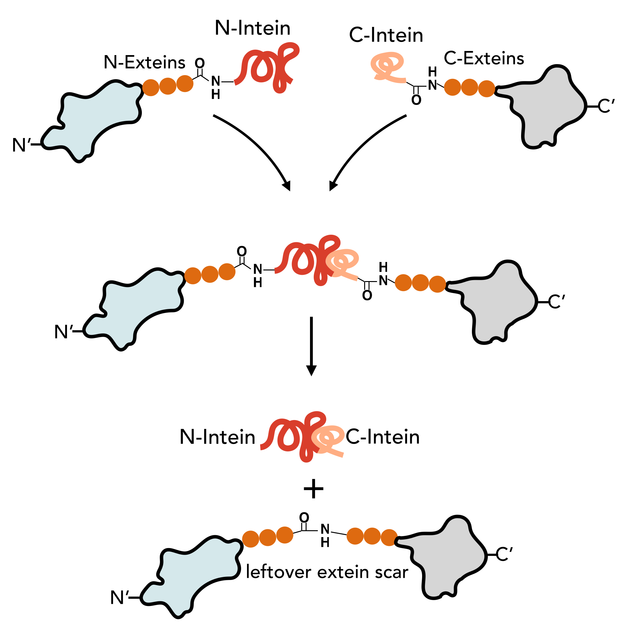 |
| Figure 1 N-intein and C-intein is spliced out by forming a leftover extein scar of the size of the N-extein and C-extein. The size and amino acid sequenceof the scar depends |
Nonribosomal peptide synthetases (NRPS) are not exclusive in their ability to produce short cyclic peptides. CyBase contains 818 entries of ribosomal head-to-tail cyclic peptides from 105 different species [1]. The peptides vary from 8 to 80 amino acids in length and in number of internal cysteine bridges that stabilizes the cyclic peptide [2]. Ribosomal cyclic peptides are potential drug candidates and their synthesis may be exploited to produce peptide drugs with increased stability or bioavailability. In one mechanism of cyclization (Figure 1), self-splicing proteins (inteins) bind and catalyze their own excision, while forming a peptide bond between the N-extein and C-extein. Numerous of inteins have been characterized, including split intein Npu DnaE from Nostoc punctiforme PCC73102 (Npu) (Figure 1) [3][4]. When the Npu DnaE intein catalyzes its own excisions, it leaves a six amino acid scar behind. The scar is called the extein. Npu DnaE has a native scar of “AEY/CFN”. This scar can in large parts be changed to other amino acids to better fit a potential synthetic peptide target [4].
Experimental Design and Methods
The BioBrick BBa_K1362000 was added by Heidelberg to the iGEM registry in 2014. It contains Npu DnaE inteins surrounding a RFP selection marker, which can be cut out with restriction enzyme BsaI. Digestion of BBa_K1362000 with BsaI generates to different overhangs, thus making it possible to insert the coding sequence (CDS) for your protein of interest. The BioBrick was used to cyclize different proteins. To our knowledge, it was not used to cyclize short peptides.
We decided to try and cyclise a synthetic 10 amino acid peptide which has similar amino acid sequence to tyrocidine. In addition, we searched the cyclic peptide database for bioactive cyclic peptide, which contained part or the entire or part of the extein scar in its native sequence. We selected a hit for a cyclotides with 33 amino acids. The sequences for both native peptides along with the sequence chosen for cyclization experiments are shown in Table 1.
| Table 1 Peptides selected for this study. Npu DnaE requires a cystine at position 1 in the extein[4]. For tyrocidine the native extein sequence was kept. | ||||||||||||
|
The peptide insert was created by designing two complementary oligos that can anneal to each other, except at their 5’ ends where they are compatible with the BsaI-generated overhangs (Table 2):
| Table 2 Oligos ordered for cycloviolacin Y1 and tyrocidine-like (see Table 1). Oligos were annealed by heating 10µM of the paired oligos to 94° C in a thermocycler. After 2 minutes the thermocycler was turned off to allow the oligos to anneal while cooling down. | ||||
|
Transformants with the cycloviolacin Y1 or tyrocidine split-intein casette were selected and grown overnight in 3mL LB media containing 6µg/mL chloramphenicol. Plasmids were purified using Qiaprep spin columns (#27104). The expression casette was combined with promoter BBa_J23101 from Anderson's promoter library. BBa_J23101 was digested with EcoRI and SpeI and the split-intein expression cassette was digested with EcoRI and XbaI for 15 minutes and subsequently heat-inactivated at 65ºC for 5 minutes. The digested expression cassette and BBa_J23101 was mixed with T4 DNA ligase in 1:3 and 1µL ligation product was transformed into chemical competent NEB DH5α E. coli cells, as previously described. Transformants were selected overnight on LB agar plates containing 6µg/mL chloramphenicol.
Expression of inteins was analyzed by SDS-PAGE using Novex NuPAGE. Samples were boiled with 1x SDS in presence of a reducing agent. The protein gel was stained using Coomassie Blue and samples were run according to the manufacture’s protocol.
Results
Transformants expressing the short peptide flanked by Npu DnaE intein (BBa_K1362000) and under control of promoter BBa_J23101 were selected and re-streaked in order to grow enough biomass for detection of peptides by chromatography. Due to time constraints, the sequence of the transformants were not confirmed. Instead, six samples for each peptide (total of 12) were analyzed by UHPLC-DAD-QTOFMS. It was not possible to detect the cyclized protein from methanol extraction of secondary metabolites. In order to confirm whether this was due to improper splicing of intein, cell lysates were analyzed by SDS-PAGE. The size of the unspliced protein iwas 19.4 kDA (cycloviolacin Y1) and 17.2 kDa (tyrocidine-like). It was not possible to detect bands at this molecular weight on the gel (data not shown). The size of the cyclic peptide was below the lower detection limit for SDS-PAGE.
Discussion
It was not possible to produce short cyclic peptides using the Npu DnaE. No unspliced variant were detected by SDS-PAGE and no cyclized peptides were detected by LC/MS. This may be explained by (1) a sequence mistake in the construct or incorrect integration of the promoter and/or (2) that the peptide was not proper cyclized. As 12 colonies were sequenced, it seems unlikely that a mutation in the oligo should be present in all transformants. The promoter may not have been inserted correctly, but the results are contradicting. Lack of cyclisation may be due to either its small size (tyrocidine) or the fact that the extein sequence was mutated from its native for expression of cycloviolacin Y1. It may also not be possible to distinguish the unspliced intein-peptide due to background in E. coli. Cyclisation of cylotides (cycloviolacin Y1 is a cyclotide) by Npu DnaE has been achieved in E. coli. [4]. It has however to our knowledge not been possible to use Npu DnaE to successfully cyclize a 10 amino acid peptide. It may be possible to achieve cyclisation of these peptides by exploring the diversity of inteins used by different organisms [5]. Heidelberg tested an intein library, which may be a start point for such intein screening in the future.
References
- Lipmann, F., Roskoski, R., Gevers, W., & Kleinkauf, H. (1970). Tyrocidine biosynthesis by three complementary fractions from Bacillus brevis (ATCC 8185). Biochemistry, 9(25), 4839–4845. doi:10.1021/bi00827a002
- Mootz, H.D., & Marahiel, M.A. (1997). The tyrocidine biosynthesis operon of Bacillus brevis: Complete nucleotide sequence and biochemical characterization of functional internal adenylation domains. Journal of Bacteriology, 179(21):6843-50
- Thorstholm L, Craik DJ. Discovery and applications of naturally occurring cyclic peptides. Drug Discov Today Technol. 2012;9:e13–e21. doi: 10.1016/j.ddtec.2011.07.005.
- Jagadish, K., Borra, R., Lacey, V., Majumder, S., Shekhtman, A., Wang, L., Camarero, J.A. Expression of fluorescent cyclotides using protein trans-splicing for easy monitoring of cyclotide-protein interactions. Angew Chem Int Ed Engl. 2013 March 11; 52(11): 3126–3131.
- Shah, N. H., & Muir, T. W. (2014). Inteins: nature’s gift to protein chemists. Chem. Sci., 5(2), 446–461. doi:10.1039/c3sc52951g
Department of Systems Biology
Søltofts Plads 221
2800 Kgs. Lyngby
Denmark
P: +45 45 25 25 25
M: dtu-igem-2015@googlegroups.com