Team:UChicago/Description
Yay, we can’t believe you fell for the clickbait. Here’s a picture of cute cats:

“Are they really going to cure sleep disorders?! How are they going to do this!?” / “sigh*……..ponders*”
Anyways, onto our project:
Background
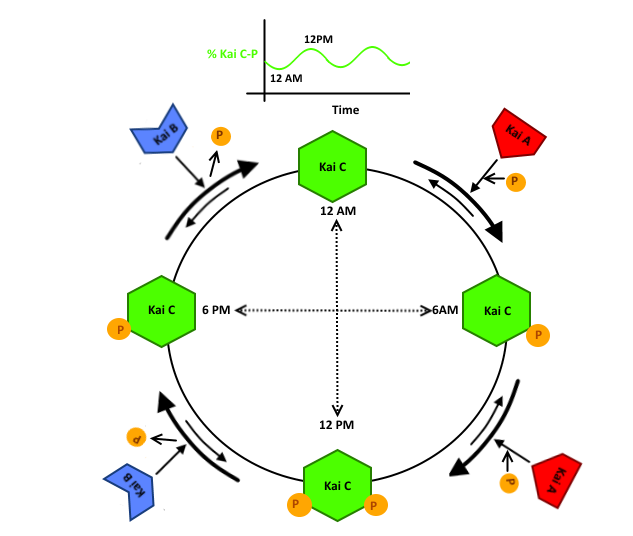
Figure 2: Schematic of how KaiB and KaiA stimulate different phosphorylation states of KaiC in a circadian rhythm. CATS.
Circadian rhythms are found in a diverse range of species, using a series of oscillatory proteins to govern everything from sleep in humans to cell cycle progression in fungi. The well characterized Kai ABC circadian oscillator in cyanobacteria has been demonstrated to regulate photosynthesis, cell division, nitrogen fixation, and a host of other essential cell processes. Because of this wide range of downstream functionality, the ability to reconstitute this robust oscillator would allow us to control the production of a target molecule on a 24 hour time scale, such as caffeine or melatonin.
The Kai ABC system is composed of three proteins, Kai A, Kai B, and Kai C. Kai A acts to autophosphorylate Kai C at one of 2 sites, either T431 (T) or S432 (S). Kai B then acts to auto-dephosphorylate Kai C. Previous research (cite that one paper)has shown that Kai B and Kai C are present in equal stoichiometries, and that the oscillatory character of the system is due to a change in Kai A concentration.
The 2015 iGem team has 3 main goals:
1) The first is to reconstitute the Kai ABC oscillator in E. Coli. The oscillator is native to cyanobacteria, and reconstituting it in the most widely used model organism in biomedical research serves as a proof of concept for further applications of the oscillatory system.
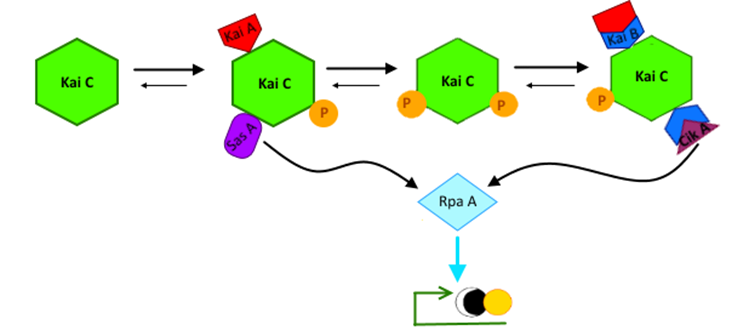
2) The second goal is to develop a readout system to allow us to assay the downstream effect of the system at various time points during its oscillation by investigating the proteins RpaA, SasA, and CikA. RpaA, SasA, and CikA are components of an expression system directly downstream of KaiABC. RpaA is a master transcription factor that connects the circadian pattern occurring on the protein-protein level in KaiABC to gene expression. SasA and CikA act in similar manner to KaiA and KaiB, phoshporylating and dephosphorylating their substrate RpaA respectively. SasA and CikA directly bind and interact with the KaiABC proteins as well as act on RpaA. While previous studies of transplanting the KaiABC system have demonstrated the ability of SasA and KaiC-P interaction to drive expression, we hope to show oscillations of the RpaA master regulator itself. With the inclusion of the negative regulator CikA, we hope to generate oscillations of RpaA and a downstream kaibc promoter which were not reported in previous studies.
3) The third goal is to link the oscillation to the expression and secretion of AANAT, an enzyme that catalyzes production of N-Acetylserotonin from tryptophan, with an eventual goal of being able to produce melatonin from tryptophan.
Melatonin Biosynthesis Pathway
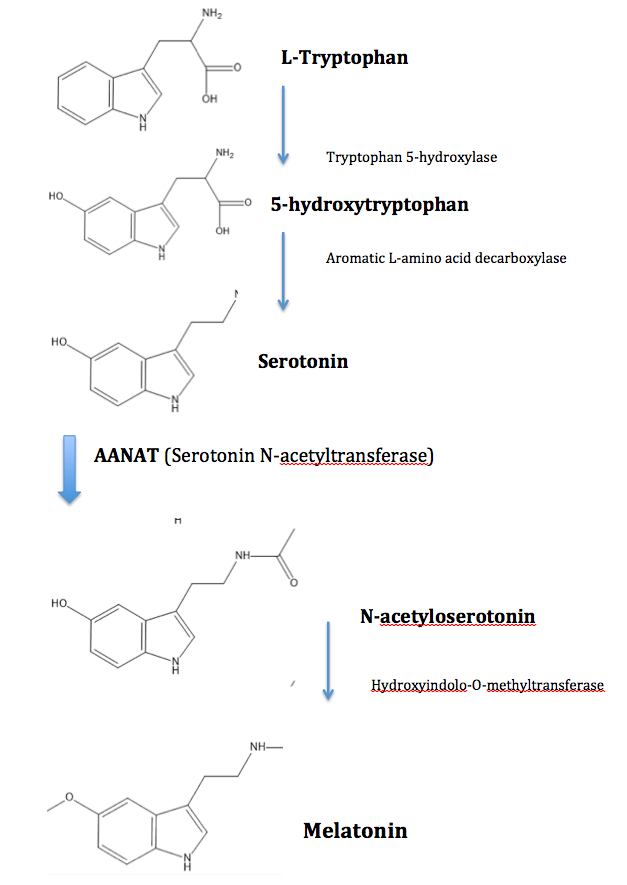
There were a couple reasons why we choose to test melatonin production control, and in turn AANAT production as our proof-of-concept project. When we were thinking of things that could be controlled with an oscillatory system, we had first though of insulin production. It seemed simple enough, we know that insulin can be produced with bacteria and people with diabetes need insulin on a timely basis, and now we have a timed delivery system to possibly deliver it in vivo. However insulin production turned out to be beyond the scope of our abilities, so we turned to sleep disorders instead, and since the sleep cycle is mainly controlled by melatonin production, we decided to focus on this. We had also considered caffeine well, however AANAT has a very low effective dosage, therefore it is more suitable for our purposes.
The biosynthesis of melatonin involves 4 steps, starting from Tryptophan, and we focused on catalyzing serotonin to N-Acetylserotonin. Not only is this step the rate-determining step, it is also how our body naturally controls melatonin production. More importantly, in the context of a gut-bacteria based drug synthesis system, N-Acetylserotonin is the first intermediate product that can pass through the blood-brain barrier, also there would be no shortage of “building blocks”, as most of the serotonin in our body is in the gut area.
Although we are focusing on the application of AANAT in sleep medicine, AANAT is also an important part of cancer research as well.
AANAT:
There were a couple reasons why we choose to test melatonin production control, and in turn AANAT production as our proof-of-concept project. When we were thinking of things that could be controlled with an oscillatory system, we had first though of insulin production. It seemed simple enough, we know that insulin can be produced with bacteria and people with diabetes need insulin on a timely basis, and now we have a timed delivery system to possibly deliver it in vivo. However insulin production turned out to be beyond the scope of our abilities, so we turned to sleep disorders instead, and since the sleep cycle is mainly controlled by melatonin production, we decided to focus on this. We had also considered caffeine well, however AANAT has a very low effective dosage, therefore it is more suitable for our purposes.
The biosynthesis of melatonin involves 4 steps, starting
from Tryptophan, and we focused on catalyzing serotonin to N-Acetylserotonin.
Not only is this step the rate-determining step, it is also how our body
naturally controls melatonin production. More importantly, in the context of a
gut-bacteria based drug synthesis system, N-Acetylserotonin is the first
intermediate product that can pass through the blood-brain barrier, also there
would be no shortage of “building blocks”, as most of the serotonin in our body
is in the gut area.
One thing that is uncertain is whether the AANAT could be secreted with endogenous secretion systems, and if they can, whether it could retain its enzyme activity. To test this out we had designed 3 constructs, unfortunately we did not have time to test them out. To learn more about them, check out our “what’s next” section in Results.
Like our team Facebook page, Genehackers@UChicago!
Questions? Comments? Send us an email!
