Team:Sydney Australia/goldengate
To assembly our multiple G-Blocks, we decided to use the Golden Gate Assembly method compared to the more traditional Gibson Assembly method and/or the classic digestion ligation method. Given this technique is in its infancy, we have provided more information below which we hope this will help future iGEM teams.
Why Golden Gate Assembly?
Golden Gate was chosen over other prominent methods as our primary cloning method as we were attempting to clone multiple inserts into the singular vector. Whilst the Gibson Assembly Method has has success in this area, given our inserts were large (upwards of 3kb) we deemed Gibson to be unsuitable for this task. Furthermore, Golden Gate only It is requires two enzymes in the reaction: DNA ligase and BsaI non-palindromic restriction enzyme.
Design
The same design protocol as prescribed in NEB website was followed.
- If there are 3 inserts (as in our project), design each end that needs to be put together to have 4bp overlapping overhangs after digestion by BsaI (as shown in above diagram).
- The left hand side overhang of the first insert and the right hand side overhang of the last insert need to overlap with the overhangs of the digested vector.
- Start by selecting the enzymes you would like to digest your vector e.g. SpeI/EcoRI and design their corresponding overhangs on the inserts to overlap with them.
- Make sure to place the BsaI recognition site outside of the ORF regions so that it is digested and removed from the final DNA e.g. place it upstream of the 5' end of insert and downstream of 3' end of insert.
- Design primers for PCR linearization of the vector such that they amplified from a few base pairs upstream of the desired cut sites other vector.
- Double check that all overhangs overlap and are in the correct overhang type of 3’ or 5’. In other words, reconstruct the final recombinant vector after drafting your designed inserts and vector to see if it yields the desired final product.
- In the thermocycling, the 37 o C cycle is for BsaI digestion and 16 o C for DNA ligase to join the overlapping overhangs together.
Method
Reaction mixture (total volume = 15 uL)
- 1 uL concentrated T4 DNA ligase (2000 U)
- 1 uL BsaI-HF (20 U)
- 1.5 uL T4 DNA ligase reaction buffer (1X)
- adjust volumes of digested vector and inserts such that they are equimolar and their total volume does not exceed 11.5 uL (50 ng of vector is an optimum amount, but there is a wide acceptable range)
- make up volume to 15 uL with MiliQ water
- mix by gentle tapping
Thermocycling
- 16 o C (10 seconds)
- 16 o C (1 minute)
- 37 o C (1 minute)
- repeat (2-3) 30-35 times
- 16 o C (1 minute)
- 50 o C (5 minutes)
- 80 o C (5 minutes)
Materials
- NEB BsaI/BsaI-HF restriction enzyme (20000 U/mL)
- NEB T4 DNA ligase concentrated (2000000 U/mL)
- NEB 10X T4 DNA ligase reaction buffer (500mM Tris-HCl, 100mM MgCl2, 10mM ATP, 100mM DTT, pH 7.5@25°C)
- Insert (equimolar amounts to vector)
- Digested PCR linearised vector (e.g. 50 ng)
Note: BsaI-HF was used in our experiments; normal BsaI should work as well.
Results & Tips
We discovered that Golden Gate gives mixed results.
- For one of our constructs, it successfully joined three 1-1.5 kbp inserted into digested pSB1C3 using the above method
- In the other construct, while the transformation gave positive results, it was discovered that instead of ligating two 2 kbp inserts into pSB1C3, it ligated a 400 bp insert in it (which is the subject of much mystery and speculation)
- Design inserts such that they contain the full ORF not divided between two inserts as this can make the design of overlapping regions complicated.
- Note that the BsaI cut sites are removed after BsaI digestion, hence, the ligated inserts will not be digested again and the BsaI will go on to digest inserts.
Overall, we believe it works well (or at least worth a try) provided that the following are implemented:
- Concentrated T4 DNA ligase
- Use a maximum total volume of 15 uL
- Use equimolar amounts of all inserts and vector (higher ratio may result in mismatched ligation)
- Make sure that all DNA samples are pure and free of contaminants (miniprep and DNA extraction kits are adequate compared to midiprep)
- Mix sample by pipetting up and down or gentle tapping
- Use 5-8 uL for transformation
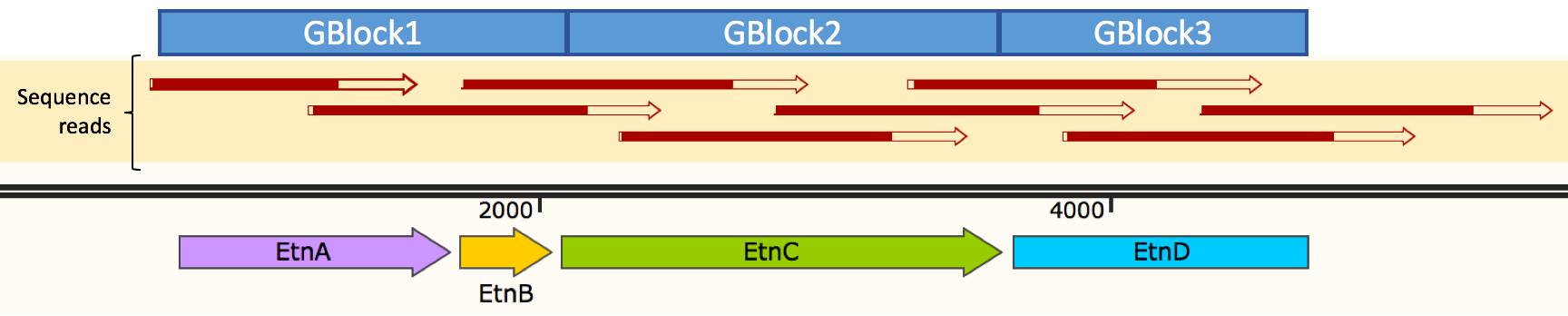
The reads demonstrate that our golden gate cloning was successful, as each GBlock was cloned in the correct orientation and order, as well showing that no mutations were introduced (100% similarity between our golden gate cloning and theoretical construct).
In conclusion, we believe that Golden Gate is a promising method and while it is at its early stages of optimisation and development, it has great potential for constructing clones with large numbers of inserts. We hope that by sharing our procedures and results we can make available more information for further optimisation and development of this method.
1 Golden Gate Assembly, New England Biolabs Inc. accessed 23/8/2015 “https://www.neb.com/applications/cloning-and-synthetic-biology/dna-assembly-and-cloning/golden-gate-assembly





