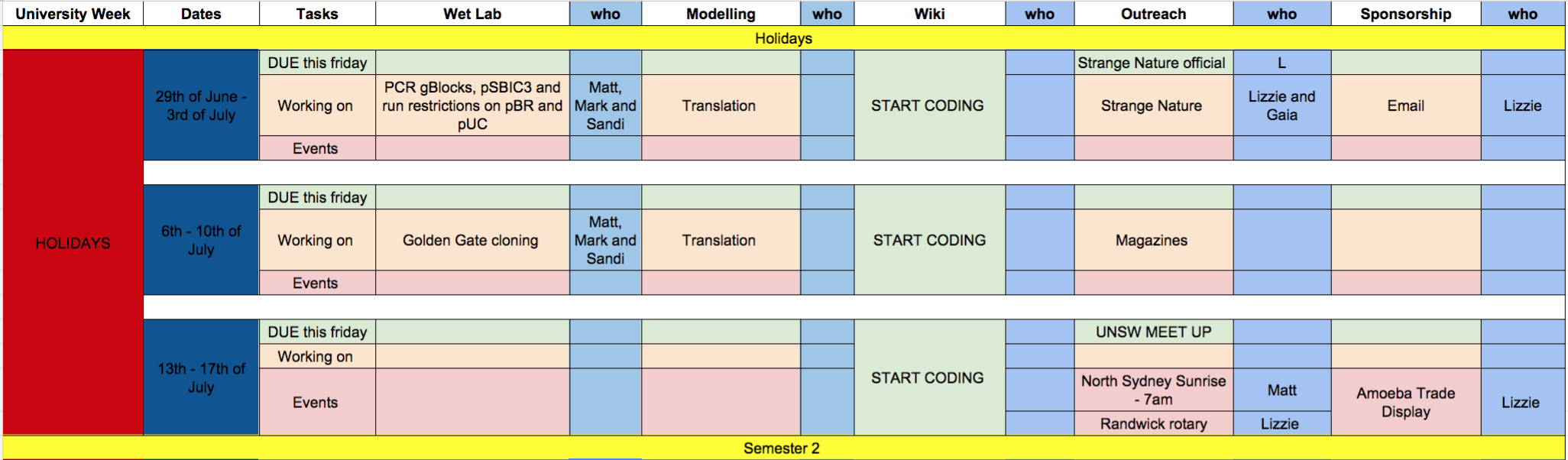
Team:Sydney Australia/Notebook
Throughout out our project we kept a detailed and through notebook and timeline. We found that having a daily, weekly, and monthly structure greatly helped in our time management. As we are a team in the Southern Hemisphere, we undertook our project during university semesters and thus effective time management was critical in helping us balance university, iGEM, work, and everything else.
Every day in the lab we would begin by asking ourselves what needed to be done that day and in what order. After every day we evaluated our progress and asked ourselves four questions -
- What did we achieve?
- How efficient were we?
- What can we improve on?
- What is the plan for tomorrow
Thus, we knew exactly what experiments needed to be done on each day. This allowed us to work faster and more efficient, it also created an easy transition between the different lab members as one member could pick up where the other left off without having to chase them down to find out the important information about the past experiment.
Furthermore, we had weekly meetings and the occassinal skype session.
"Progress is made by trial and failure; the failures are generally a hundred times more numerous than the successes ; yet they are usually left unchronicled"
This was not the case for our team as we sought to extensively chronicle all the successes and failures. We hope our observations and troubleshooting will help future iGEM teams.
Wet Lab
Tuesday
- Meet up for the first time and were intrdouced to the project. Started to do some research on the project
Modelling
Thursday-Friday
- Researched the possibility of developing a tool for simulation of the translation process.
- Harry & Matt discussed this possibility, along with potential areas for improvement.
Wet Lab
Friday - Sunday
- Underwent necessary safety training to work independently in the PC2 laboratory
Human Practices
Monday - Thursday
- Started to brainstorm ideas for a human practices project and outreach activities
Modelling
Wednesday
- After continued research over the weekend, met with academic staff in the department to discuss sensible mechanisms that should be included in simulating, and potentially optimising, translation.
Thursday-Friday
- Continued research, focusing on the work of Brakley et al, as well as different aspects of our organisms of interest (and how they compare to yeast).
Wet Lab
Started designing the G-Blocks for order and began creating electrocompetent cells and various agar plates
Human Practices
Tuesday - Thursday
- Developed the Strange Nature question and created the information packages and contacted Rotary clubs
Modelling
Wednesday - Friday
- Begin to develop a translation simulation tool in MATLAB.
- Continued review of literature to find appropriate modelling parameters.
Wet Lab
Continued to work on the G-Blocks with the model created
Human Practices
Friday - Saturday
- Developed and worked on the Rotary Presentations
Modelling
Monday - Thursday
- Re-thougth the coding approach being taken, as it may need to be scaled up to treat larger problems in the future. To achieve this we made maximum use of MATLAB's vectorisation to speed up execution.
Wet Lab
Continued to research and plan the design of the G blocks. Discovered the world of Golden Gate cloning and decided to apply it to our project
Modelling
Thursday - Sunday
- Continued to develop aspects of our visualisation tool, now focusing on how we can best display the simulated processes, to give the user some idea of what it is revealing about the cell.
Wet Lab
Monday - Friday
- Ordered the G-Blocks and waited for them to arrive
Human Practices
Wednesday
- Discover Uni Day at Sydney University. Our first outreach event went very well!
Modelling
Tuesday
- With the visualisation tool now working, other members of the team test it. This is to improve usability and to reveal any flaws in its operation. A number of suggestions were taken for future improvement
Friday
- Made changes to the program to reflect advice received from all members of the team.
Wet Lab
Wednesday and Thursday
- Aim - To replicate the findings of Mai Anh Ly's PhD thesis that showed low levels of expression of the EtnABCD cluster in Pseudomonas Putida KT2440
- Result - Confirmed the findings
Friday
- Aim - To transform the pBP-Etn plasmid into pre-made componenet Pseudomonas Putida cells by electroporation
- Results - Successful transformation
Human Practices
Monday - Thursday
- Began research on the business proposal and sought contacts in the industry
Modelling
Monday-Tuesday
- Continued testing of visualisation tool, and validation of its capability to demonstrate a wide range of anticipated behaviours.
Wednesday - Friday
- Begun researching translation optimisation processes. A number exist in the literature, as well as via online tools provided by biotech companies.
Wet Lab
Monday
- Aim - Assess if the cells transformed with the wild type EtnABCD cluster have any monooxygenase activity through a Nitrobenzylpyridine assay
- Result - Positive test had low (but notable) absorbance compared to control.
Human Practices
Thursday - Saturday
- Continued to work on the business plan and began collating a list of schools to contact for Strange Nature
Modelling
Monday - Thursday
- Continued research of mRNA translation and the factors which effect its performance.
- Read literature regarding how different aspects of mRNA sequences are believed to cause certain behaviours during the translation process.
Friday
- Discussed the possibility of making a device which would automatically control electrophoresis experiments.
Human Practices
Monday
- Started contacting the media
Modelling
Monday - Tuesday
- Decided to build a device which could control electrophoresis experiments, came up with a preliminary circuit design.
Tuesday - Friday
- Continued to research translation optimisation processes, as well as potentially mathematical approaches to achieve these.
- Noticed the work of Spencer et al which presents a new (and so far successful) methodology for predicting translation rates of different codons.
Friday
- Order initial components for electrophoresis device
Wet Lab
Monday
- Aim - First PCR of the ordered G-Blocks.
- Result - Unsuccessful. High molecular weight smears were present
Human Practices
Friday - Sunday
- Began to write and edit articles for the papers that had agreeded to publish articles
Modelling
Tuesday - Thursday
- Continual research on the work of Spencer et al, as well as investigation of future research which make reference to the initial 2012 paper.
Wet Lab
Exam time. We took a break from iGEM
- Implemented the standard codon harmonisation methodology using Spencer et al quantisation of rates. This method is based on that proposed by Kudla et al, which has found widespread use, though it makes a wide range of assumptions when quantising translation rates.
Modelling
Wednesday - Saturday
Modelling
Monday - Wednesday
- Testing of the standard codon harmonisation software, comparing results with those produced by various online tools which claim to do the same thing.
Thursday
- Electronic components for the electrophoresis device arrive by mail.
Human Practices
Thursday
- Briefly worked on the business plan
Modelling
Monday - Friday
- Prototype the circuit for use with the electrophoresis device.
- Develop code for the device, which by Thursday is working correctly.
Wednesday - Friday
- Continue investigating factors important in mRNA translation.
- Research simulated annealing, a statistical approach to optimisation which may be used.
Wet Lab
Monday
- Aim - PCR amplification of pBSIC3 using Phusion and visualization using gel electrophoresis
- Result - Several bands present, yet bigger/smaller than expected 2 kb fragment
Tuesday
- Aim - A gel was run with a thermocycling gradient (59.4, 61.3, 63.6, 66.2, 69, 71.6, 74, 76) to determine the best annealing temperature.
- Result - The thermocycling gradient showed that 69 was the most suitable temperature.
- Aim - PCR amplification (annealing temperature of 69 and 98 cycles) and gel electrophoresis of EtnAB, EtnC, EtnD, Gro1, Gro2 G-blocks using primers iGEM1501 and iGEM1502. The gel was stained with Gel-Red.
- Result - The gel electrophoresis showed high molecular weight smears. These were though to be the result of contaminants in the reagent.
Wednesday
- Aim - Second attempt of PCR amplification and gel electrophoresis of pBSIC3 with an annealing temperature of 69 and 35 cycles instead of 98
- Result - The gel showed no DNA bands and no smears for all lanes.
- Aim - DpnI treatment of pSB1C3 PCR sample (restriction, transformation and column purification)
- Result - successful elimination of plasmid using DpnI
Thursday
- Aim - Make up new working stock of primers 1501 and 1502 and the third attempt at running the PCR and gel electrophoresis of pBSIC3 from 30/6/15 and 1/7/15.
- Result - The no-template-control yielded no DNA bands. All other lanes showed smears.
Friday
- Aim - Fourth attempt at PCR amplification and gel electrophoresis of pBSIC3 from the 30/6/15, 1/7/15 and 2/7/15.
- Changes to the PCR protocol *same conditions for Gro2 *GC buffer instead of HF buffer for Gro2 *Using Taq pol and Taq buffer for Gro2 *1/10 of DNA template *Shorter primers *Same conditions using Abi's G blocks
- Results - Large band of correct size was seen with Taq polymerase, using the same primer. Both G-blocks and the primers are working as expected. Next step: eliminate all plasmid, digestions with more DpnI should be performed again with more DpnI
Human Practices
Sunday
- Began developing the activities for APCS
Modelling
Monday
- Test the prototype electrophoresis circuit, now named ElectroStop
- ElectroStop demonstrates a capability, after some tuning, to be able to accurately sense the presence of marker dye obstructing its laser.
Tuesday - Thursday
- Adapt the standard codon harmonisation tool to accurately reflect the rate quantisations proposed by Spencer et al, which seems more physically realistic.
Thursday - Friday
- Begin coding a new codon optimisation tool, now named TransOpt, which we hope will optimise many aspects of the translation process.
Wet Lab
Monday
- Aim - Testing PCR reactions of gBlocks against varying polymerase activities
- Results - Full strength Phusion and Pfu all seemed to produce strong bands
Tuesday
- Aim - PCR of monooxygenase G blocks
- Results - Smears and bands of incorrect sizes were observed
- Aim - PCR amplification of G blocks with Pfu polymerase
- Results - Smears and bands of incorrect sizes were observed
Wednesday
- Aim - Sixth attempt of PCR amplification and gel electrophoresis of EtnAB, EtnC, and EtnD
- Results - All G blocks are pure, bands are evident
- Aim - Golden Gate assembly of GroEl/ES to pSBIC3 PCR product
- Results - No colonies appeared after 20 hours. Colonies appeared in JM109 transformed with GFG of pSBIC3-GroEL/ES
Thursday
- Aim - Repeat PCR from 8/7/15 with a new set of pipettes
- Results - Multiple products observed for all lanes including the NTC
- Aim - Amplification of pSB1C3 with Phusion
- Results - Large clear band was observed around 1/5kb which is the expected size of pCB1C3
- Aim - Golden Gate Assembly of GroEL/ES and pUCP24
- Results - Cloning was not successful
Human Practices
Monday - Thursday
- Tested out the gels for the APCS and began to create bacterial paintings which could be used at APCS
Modelling
Monday - Wednesday
- Continue developing the code for TransOpt. Now try to make reading of organisms data (e.g. Gene Copy Numbers) as general as possible, such that data from new organisms can be introduced in the future.
Thursday - Sunday
- Design structure for ElectroStop in Solidworks, which will then be 3D Printed.
- Order some new electrical components which earlier testing indicated a need for.
Wet Lab
Friday
- Aim - Amplification of AE-blue-aac with the primer set used in pervious reactions and a control primer set (igem1501 and igem1502) to determine whether contamination is in the primer
- Results - Band of correct size observed for both PCR reactions but there were still smears in all lanes. No difference between control and test PCRs. Cause of smears is unknown
Saturday
- Box PCR for outreach activities
Human Practices
Tuesday - Friday
- Started to wrok on the Strange Nature website and finalised the question. Officially launched Strange Nature
Modelling
Monday
- Testing the visualisation tools: They appear to show scientifically-reasonable results which agree with what we had expected.
Tuesday
- Finalise the design of the ElectroStop structure in Solidworks, ensure measurements match up with all common electrophoresis trays in our lab.
Wednesday - Friday
- Continue development of TransOpt, now designing an energy function for use in the simulated annealing approach.
Wet Lab
Tuesday
- Aim - PCR amplification of GroEL/ES and pSB1C3 and gel visualisation
- Result - Smears were observed on the gel
Wednesday
- Aim - PCR amplification of GroEL/ES with Phusion
- Result - Smears were observed on the gel. Next approach was to try the Pfu
Thursday
- Aim - PCR amplification of GroEL/ES with Pfu
- Result - Smears were observed on the gel
- Aim - pSB1C3/pUCP24 digestion for use of GG-cloning with Gro
- Result - Smears were observed on the gel
Friday - Saturday
- Aim - Digest of pSB1C3 with SpeI/EcoRI and golden gate cloning of pSB1C3/Gro
- Result - Colonies appeared which need to be screended and inoculated
Human Practices
Monday
- We attended APCS and spoke to the students about iGEM, synthetic biology, and our project
Tuesday
- We painted on plates with the chromoproteins from the Uppsala team for outreach activities
Modelling
Monday
- Use the Sydney Invention Studio to 3D Print the structure for ElectroStop.
- After four hours printing, structural members are finished in VeroWhite plastic, and appear to be manufactured as per design.
Wednesday-Sunday
- Continue development of TransOpt, now focusing on how randomised changes are kept / rejected by the optimisation algorithm.
- Extend TransOpt to work with data files from other organisms.
- TransOpt's core capabilities finished and working for all organisms we will use in our project.
- Utilise TransOpt to produce EtnABCD Sequences for use in main project.
Wet Lab
Monday
- Aim - Patch and left junction PCR screening of GroEL/ES transformants followed by gel visualisation
- Results - 14 colonies suspected of carrying successful clones, 3 colonies showed clear bands in gel
Friday
- Aim - Colony spanning PCR putative GroEL/ES clones
- Results - Cloning was unsuccessful
Modelling
Monday
- New electrical components arrive for ElectroStop.
- Testing of new components is successful.
Wednesday - Sunday
- Begin testing TransOpt with a variety of input sequences.
- Appears to provide sensible answers, though convergence (due to inefficient programming) is at this stage slow.
- Utilise TransOpt to produce fluorescent protein sequences for use in experiments, both via standard codon harmonisation, fast codon utilisation, and the TransOpt method.
Friday
- Solder all electrical components to ElectroStop's protoboard, as prototyping and testing has revealed that the current design functions as necessary.
Wet Lab
Monday
- Aim - Right-junction colony PCR of pSB1C3-Gro JM109 clones
- Results - No bands in PCR gel visualisation, therefore the cloning was unsuccessful
Wednesday
- Aim - Digest of pSB1C3 with EcoRI and SpeI to prepare for a second attempt at GG cloning with GroES/EL G blocks
- Results - Bands visualized on gel, therefore successful
Thursday
- Aim - GG cloning of pSB1C3, GroEL/ES and transformation
- Results - some test colonies were present, so cloning looked to be somewhat successful
Friday
- Aim - colony PCR (left-junction) of pSB1C3-Gro GG clones from yesterday and visualisation
- Results - successful: bands as expected
Human Practices
Monday
- Began to look into the LacI co-expression as a solution to the toxicity encountered when having to produce this on an industrial scale
Modelling
Monday - Thursday
- Continue testing of TransOpt, experiment with different values of alpha and beta values to determine how they affect the final outputted result.
Thursday - Friday
- Assemble circuit and 3D printed components to create final prototype of ElectroStop.
- Test the ElectroStop device on sample dyes implanted in a gel.
- Works effectively on a range of dyes, though purple-tinted ones provide the greatest sensitivity.
Wet Lab
Monday
- Aim - spanning colony PCR and broth inoculation of putative GroES/EL clones to confirm assembly and gel visualisation
- Result - bands not expected size, therefore deemed inconclusive
Tuesday
- Aim - right junction colony PCR of putative GroEL/ES clones
- Result - expected band seen, however as spanning PCR was unsuccessful, cannot confirm Gro assembly.
Wednesday
- Aim - pSB1C3-EtnABCD GG cloning and transformation
- Result - some successful colonies
Thursday
- Aim - spanning colony PCR of putative EtnABCD clones to confirm assembly and gel visualisation
- Result - no bands shown in white (putative) colonies so gel inconclusive
Human Practices
Friday
- Lizzie attended Kambala School and spoke to the girls about iGEM, synthetic biology, and our project.
Saturday
- We attended Science in the Swamp and spoke to over 10 000 about synthetic biology and our project
Modelling
Monday - Thursday
- Continue testing of TransOpt.
- Add capability for removal of iGEM restricted sites from the final result, and during the optimisation process.
Wednesday - Friday
- Test ElectroStop with gel runs to ensure switching capability is sufficient.
- Success - Appears able to regularly switch the 120-180 V power supplied to the electrophoresis tray.
Wet Lab
Monday - Wednesday
- Aim - inoculation, plasmid purification, digestion and visualisation of EtnABCD-pSB1C3 GG cloning
- Result - 4/5 clones were identified as positive
Thursday
- Aim - digest using EcoRI and SpeI to confirm insertion of GroES/EL construct followed by gel visualisation
- Result - not promising; appears a 400bp fragment inserted
Friday
- Aim - EcoRI-SpeI digestion of EtnABCD-pSB1C3 and pBBR-MCS2
- Result - mix of white, green and mixed green/white large colonies observed
Human Practices
Saturday
- We had a stall at the University of Sydney Open Day and spoke to prospective students about iGEM and out sompetition
Modelling
Tuesday - Friday
- Continue testing of TransOpt
- Add capability for graphic results as they are generated.
- Add capability for converting output back to base letters (A,T,C,G) and storing in an easily retrievable text file.
- Add final check after optimisation is complete, to ensure that generated sequence is synonymous to original sequence.
Wet Lab
Wednesday
- Aim - plasmid purification, digest, gel visualization of EtnABCD-pBBR with EcoRI & EcoRI+SpeI
- Result - some clones showed the correct insert
- Aim - transformation of positive EtnABCD-pBBR clones into Pseudomonas putida
- Result - no test colonies were observed so transformation deemed unsuccessful
Thursday - Friday
- Aim - digestion and ligation of E. coli genomic GroEL/ES into pUCP24/pUS44/pSB1C3 and transformation into JM109
- Result - all of these ligations were successful
Wednesday - Thursday
- Aim - NBP assay to investigate monooxygenase activity in putative clones & replicate
- Result - no change in absorbance detected, thus unsuccessful
- Aim - digestion of pUS44-lacI with SalI, ligation and transformation into E. coli
- Result - some colonies on test plate, therefore successful
Human Practices
Wednesday
- We had a stall at the Investig8 Uni Day and taught the students about synthetic biology and our project
Modelling
Monday - Tuesday
- Collect final results from visualisation tool and form videos / graphs for use on Wiki.
- Use visualisation tool on some sequences generated by TransOpt.
- Compiling final software for visualisation tool.
Tuesday - Thursday
- Complete final testing of TransOpt.
- Begin creating a simplified final version for upload.
Tuesday - Friday
- Experimentally deploy ElectroStop device for usage.
Wet Lab
Monday
- Aim - preparing pSB1C3-EtnACBD clones for sequencing efforts
- Results - yield and purity were sufficient to go ahead
- Aim - PCR screening of putative pSB1C3-lacI clones using left/right junction primers and gel visualisation Results - a number of potential insert-containing clones. Go on to incoulate
Tuesday
- Aim - isolate, restriction digest and gel visualization of pSB-lacI clones to confirm they contain insert
- Results - one clone had correct insert
Wednesday
- Aim - transformation of pSB-lacI clones into chemically competent E. coli JM109-pBBR1MCS2 cells
- Results - yesterday’s successful clone transformed and plated; odd growth on different selective media
Thursday
- Aim - transformation of pBBR-Etn into new electrocompetent Pseudomonas cells
- Results - unsuccessful
Friday
- Aim - pSB1C3-Etn and pBBRMCS2 digestion, ligation and transformation into Pseudo K2440 for Etn-pBBR
- Results - two clones showed good growth
Saturday
- Aim - transformation of Pseudo KT440 with lacI-pUS44
- Results - a few dozen colonies of pUS44-lacI
Human Practices
Friday
- Contacted more papers
Modelling
Monday - Wednesday
- Final compilation of TransOpt script for upload to Wiki.
- Analysis of TransOpt experimental on fluorescent protein.
Thursday
- Use ElectroStop in lab.
Wet Lab
Tuesday
- Aim - spanning colony PCR of colonies from Saturday’s pUS44-lacI clones
- Results - showed correct insert
- Aim - Pseudo-pUS44-lacI competent cells and colony PCR of successful colonies
- Results - 3 colonies showed to be successful
Thursday
- Aim - spanning colony PCR of colonies from Saturday’s pUS44-lacI clones
- Results - a dark purple colour was produced, indicating a positive result
- '''Aim - parts submission (lacI, BsFP, etnABCD all in pSB1C3)'''
- '''Results - SUCCESS!'''
Modelling
Monday - Friday
- Finished analysing experimental results from fluroprotein: Found that TransOpt optimised sequence showed very poor fluroescence.
- Discuss and try to hypothesise causes for TransOpt sub-par performance on this particular protein