Difference between revisions of "Team:Goettingen/Description"
| Line 37: | Line 37: | ||
<p><strong>Phosphatase</strong></p> | <p><strong>Phosphatase</strong></p> | ||
<p><strong>Castillo Villamizar, G., Nacke, H., Böhning, M., Elisabeth, G., Kaiser, K., Daniel, R.</strong> (2014) Novel phosphatases derived from functional metagenomics. 7<sup>th</sup> International Congress on Biocatalysis. <em>Biocat2014 Hamburg. </em>August 32 –September 4, 2014, P. 87</p> | <p><strong>Castillo Villamizar, G., Nacke, H., Böhning, M., Elisabeth, G., Kaiser, K., Daniel, R.</strong> (2014) Novel phosphatases derived from functional metagenomics. 7<sup>th</sup> International Congress on Biocatalysis. <em>Biocat2014 Hamburg. </em>August 32 –September 4, 2014, P. 87</p> | ||
| + | |||
| + | <p><strong>Greiner, R., Farouk, A.-E. </strong>(2007) <a href=" http://link.springer.com/article/10.1007/s10930-007-9086-z"> Purification and Characterization of a Bacterial Phytase Whose Properties Make it Exceptionally Useful as a Feed Supplement.</a>. <em>The Protein Journal. </em>Volume 26, Issue 7, Pp. 467-474</p> | ||
| + | <p> </p> | ||
Revision as of 19:52, 18 September 2015

About our Project
Our project
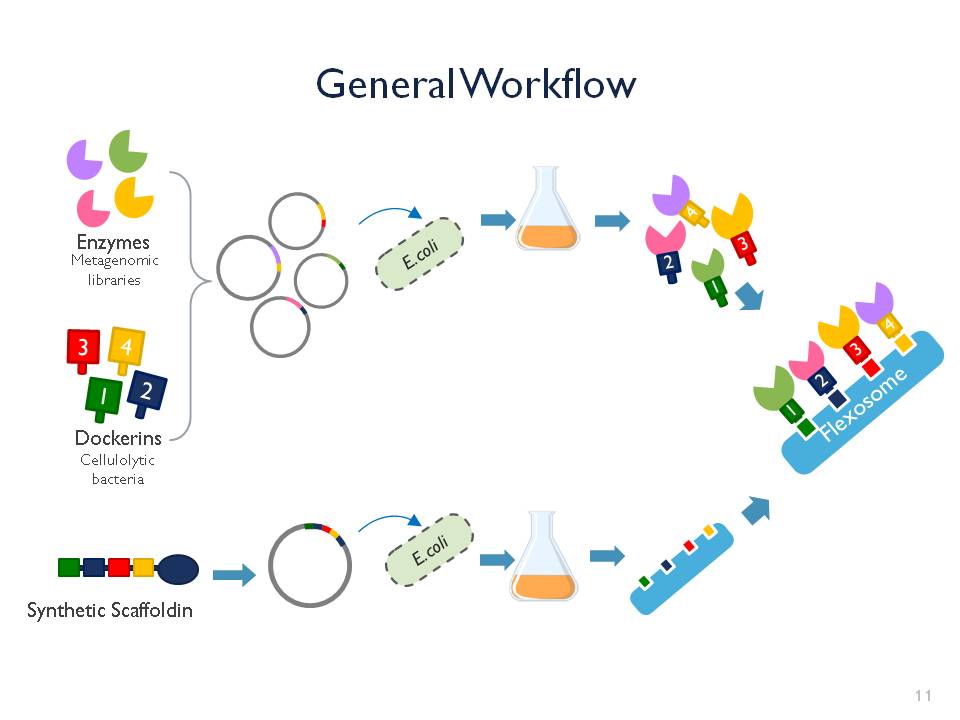
The iGEM team Goettingen 2015 is currently developing the “Flexosome”. It is our enzymatic penknife: a customizable complex for more efficient and synergistic multi-enzymatic processes. It consists of a scaffoldin, dockerins and exchangeable enzymes. The enzymes of interest can be attached to the scaffoldin via the dockerin stations and once attached complete the reactions.The first step
Right now the team is working on a proof of concept. Our first Flexosome will contain phosphatase, esterase and cellulase found in metagenomic libraries along with RFP, a fluorescent reporter gene. Once our E. coli is able to express and produce this functional Flexosome, this will open the door for more complex and specific applications.The inspiration
We studied the natural cellulosome structure, in which different cellulotytic enzymes are assembled into a bigger scaffoldin via dockerin stations, leading to optimized cellulose degradation. The interaction between scaffoldin and dockerins has been proven to be strong and stable, this gave us the ideal platform to design our "Flexosome” device. Traditionally cellulosome engineering is thought for optimization of cellulose degradation, but we don’t have to stop there! A much broader range of enzymes are actively being discovered every year. That is why we intend to combine the scaffoldin protein from cellulolytic bacterium with varying and exchangeable enzymes. The result: a very versatile, stable and innovative tool!The goal
We want to achieve a protein construct which will be able to ensure a high local concentration of enzymes and catalyse multiple enzymatic processes at one site, making the product of one process the substrate of the next on the scaffoldin. Ultimately, the construct will not only increase efficiency of the reactions but also be fully customisable for any kind of enzymatic process.
References
Esterase
Nacke, H., Will, C., Herzog, S., Nowka, B., Engelhaupt, M., Daniel, R. (2011) Identification of novel lipolytic genes and gene families by screening of metagenomic libraries derived from soil samples of the German Biodiversity Exploratories. FEMS Microbiology Ecology. Volume 78, Issue 1, Pp. 188 - 201
Phosphatase
Castillo Villamizar, G., Nacke, H., Böhning, M., Elisabeth, G., Kaiser, K., Daniel, R. (2014) Novel phosphatases derived from functional metagenomics. 7th International Congress on Biocatalysis. Biocat2014 Hamburg. August 32 –September 4, 2014, P. 87
Greiner, R., Farouk, A.-E. (2007) Purification and Characterization of a Bacterial Phytase Whose Properties Make it Exceptionally Useful as a Feed Supplement.. The Protein Journal. Volume 26, Issue 7, Pp. 467-474