Difference between revisions of "Team:Goettingen/Experiments"
| (102 intermediate revisions by 7 users not shown) | |||
| Line 1: | Line 1: | ||
{{Goettingen}} | {{Goettingen}} | ||
<html> | <html> | ||
| − | |||
<!-- Start of CSS--> | <!-- Start of CSS--> | ||
<style type="text/css"> | <style type="text/css"> | ||
| − | h1 { | + | #contentContainer h1 { |
color:white; !important; | color:white; !important; | ||
text-align:center; | text-align:center; | ||
| Line 15: | Line 14: | ||
padding:5px; | padding:5px; | ||
line-height: 1.5; | line-height: 1.5; | ||
| − | } | + | font-size:small; |
| + | transition: background 0.8s; | ||
| + | -moz-transition: background 0.8s; | ||
| + | -webkit-transition: background 0.8s; | ||
| + | -o-transition: background 0.8s; | ||
| + | -ms-transition: background 0.8s; | ||
| + | } | ||
| + | |||
| + | #contentContainer h1:hover { | ||
| + | color:white; | ||
| + | background-color:#7fbbe4; | ||
| + | transition: background 0.8s; | ||
| + | -moz-transition: background 0.8s; | ||
| + | -webkit-transition: background 0.8s; | ||
| + | -o-transition: background 0.8s; | ||
| + | -ms-transition: background 0.8s; | ||
| + | } | ||
a { | a { | ||
| Line 22: | Line 37: | ||
} | } | ||
| − | #menu1, #menu2, #menu3, #menu4, #menu5, #menu6, #menu7, #menu8, #menu9, #menu10, #menu11, #menu12{ | + | #menu1, #menu2, #menu3, #menu4, #menu5, #menu6, #menu7, #menu8, #menu9, #menu10, #menu11, #menu12, #menu13, #menu14, #menu15, #menu16, #menu17, #menu18, #menu19, #menu20, #menu21, #menu22, #menu23, #menu24, #menu25, #menu26, #menu27, #menu28, #menu29, #menu30, #menu31, #menu32, #menu33, #menu34, #menu35, #menu36, #menu37, #menu38, #menu39, #menu40, #menu41{ |
display:none; | display:none; | ||
width:90%; | width:90%; | ||
| Line 40: | Line 55: | ||
</style> | </style> | ||
<!-- End of CSS --> | <!-- End of CSS --> | ||
| − | + | <h2> Media/Buffer </h2> | |
| − | <a href="" onClick=" $('#menu1').slideToggle(300, function callback() { }); return false;"><h1>LB Medium</h1></a> | + | <a href="" onClick=" $('#menu1').slideToggle(300, function callback() { }); return false;"><h1 style="color:white;">LB Medium</h1></a> |
<div id="menu1"> | <div id="menu1"> | ||
| Line 76: | Line 91: | ||
</td> | </td> | ||
<td valign="top" width="150"> | <td valign="top" width="150"> | ||
| − | <p> | + | <p>ad. 1000 mL</p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 85: | Line 100: | ||
</ul> | </ul> | ||
<p>- Autoclave at 121 <sup>o</sup>C for 20 min.</p> | <p>- Autoclave at 121 <sup>o</sup>C for 20 min.</p> | ||
| − | <p>- The preferred antibiotic is added | + | <p>- The preferred antibiotic is added from a concentrated stock solution to an appropriate concentration (e.g Ampicilin is added to a final concentration of 100 µg/ml).</p> |
<ul> | <ul> | ||
<li>To the liquid medium antibiotic is added upon usage.</li> | <li>To the liquid medium antibiotic is added upon usage.</li> | ||
| Line 92: | Line 107: | ||
</div> | </div> | ||
| + | <a href="" onClick=" $('#menu28').slideToggle(300, function callback() { }); return false;"> | ||
| + | <h1>"Fat" LB Medium</h1></a> | ||
| + | <div id="menu28"> | ||
| + | <p> Fat medium was used as culture medium for the induction of our cultures </p> | ||
| + | <table border="1" cellpadding="0" cellspacing="0"> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td valign="top" width="307"> | ||
| + | <p> | ||
| + | Yeast extract | ||
| + | </p> | ||
| + | </td> | ||
| + | <td valign="top" width="97"> | ||
| + | <p> | ||
| + | 40 g | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="307"> | ||
| + | <p> | ||
| + | KH<sub>2</sub>PO<sub>4</sub> (MW: 136.09) | ||
| + | </p> | ||
| + | </td> | ||
| + | <td valign="top" width="97"> | ||
| + | <p> | ||
| + | 7.0 g | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="307"> | ||
| + | <p> | ||
| + | Na<sub>2</sub>HPO<sub>4</sub> x 12 H<sub>2</sub>O (MW: 358.14) | ||
| + | </p> | ||
| + | </td> | ||
| + | <td valign="top" width="97"> | ||
| + | <p> | ||
| + | 15.14 g | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="307"> | ||
| + | <p> | ||
| + | H<sub>2</sub>O dest. | ||
| + | </p> | ||
| + | </td> | ||
| + | <td valign="top" width="97"> | ||
| + | <p> | ||
| + | ad. 1000 mL | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | <p> | ||
| + | pH 6.8 with NaOH (ca. 10mL 1M NaOH) | ||
| + | </p> | ||
| + | <p> | ||
| + | After autoclaving add: | ||
| + | </p> | ||
| + | <table border="1" cellpadding="0" cellspacing="0"> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td valign="top" width="307"> | ||
| + | <p> | ||
| + | 50% Glycerine (sterile) | ||
| + | </p> | ||
| + | </td> | ||
| + | <td valign="top" width="97"> | ||
| + | <p> | ||
| + | 14 mL | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="307"> | ||
| + | <p> | ||
| + | 24% MgSO<sub>4</sub> x 7 H<sub>2</sub>O (sterile) | ||
| + | </p> | ||
| + | </td> | ||
| + | <td valign="top" width="97"> | ||
| + | <p> | ||
| + | 7.5 mL | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | </div> | ||
| + | <a href="" onClick=" $('#menu37').slideToggle(300, function callback() { }); return false;"><h1 style="color:white;"> Media and Culture Methods for Dockerin Organisms of Origin</h1></a> | ||
| + | <div id="menu37"> | ||
| + | <p> | ||
| + | <strong>Pseudobacteroides cellulosolvens</strong> | ||
| + | </p> | ||
| + | <p> | ||
| + | <a href="https://www.dsmz.de/microorganisms/medium/pdf/DSMZ_Medium315.pdf">DSMZ Medium 315</a> | ||
| + | </p> | ||
| + | <p> | ||
| + | <strong>Clostridium cellulolyticum</strong> | ||
| + | </p> | ||
| + | <p> | ||
| + | <a href="https://www.dsmz.de/microorganisms/medium/pdf/DSMZ_Medium520.pdf"> DSMZ Medium 520</a> | ||
| + | </p> | ||
| + | <p> | ||
| + | <strong>Clostridium thermocellum</strong> | ||
| + | </p> | ||
| + | <p> | ||
| + | <a href="https://www.dsmz.de/microorganisms/medium/pdf/DSMZ_Medium122.pdf">DSMZ Medium 122</a> | ||
| + | </p> | ||
| + | <p> | ||
| + | <strong>Acetivibrio cellulolyticus</strong> | ||
| + | </p> | ||
| + | <p> | ||
| + | <a href="https://www.dsmz.de/microorganisms/medium/pdf/DSMZ_Medium165.pdf">DSMZ Medium 165</a> | ||
| + | </p> | ||
| + | <p> | ||
| + | <a href="http://www.dsmz.de/microorganisms/medium/pdf/DSMZ_Medium141.pdf">DSMZ Medium 141</a> | ||
| + | </p> | ||
| − | + | </div> | |
| − | <div id=" | + | <a href="" onClick=" $('#menu15').slideToggle(300, function callback() { }); return false;"> |
| − | + | <h1>Phosphatase Activity plates, Sperber media</h1></a> | |
| − | + | <div id="menu15"> | |
| − | + | <p style="text-align: center;"><span style="font-size: small; color: #888888;"><strong></strong></span></p> | |
| − | + | <p style="text-align: center;"><span style="font-size: x-large; color: #888888;"><strong></strong></span></p> | |
| − | + | <p> </p> | |
| − | + | <p><span style="text-decoration: underline;">A: Stock reagents</span></p> | |
| − | + | <p>1M IPTG (4.76 g in 20ml Millipore-H2O) filter with blue 0.22 µm sterile filter and freeze at -20˚C</p> | |
| − | + | <p>50mg/ml Kanamycin in Millipore H2O, filter with blue 0.22 µm sterile filter and freeze at -20˚C</p> | |
| − | + | <p>100mg/ml Ampicillin in 50% ethanol, filter with blue 0.22 µm sterile filter and freeze at -20˚C</p> | |
| − | + | <p>25mg/ml BCIP (in DMF) filter with blue 0.22 µm sterile filter and keep in a falcon tube wrapped with aluminum foil at 4˚C. BCIP: Biomol Nr. 2291 (MG 433.64)</p> | |
| − | + | <p>Glycerol (99%), autoclaved and kept at room temperature</p> | |
| − | + | <p> </p> | |
| − | + | <p><span style="text-decoration: underline;">B: Sperber medium</span></p> | |
| − | + | <p>1g Yeast extract</p> | |
| − | + | <p>3.5 ml 50% w/w Phytic acid</p> | |
| − | + | <p>0.2 g CaCl2</p> | |
| − | + | <p>0.5 g MgSO4</p> | |
| − | + | <p>Adjust the pH to 7.2 with NaOH</p> | |
| − | + | <p>Ad 2 L of Millipore H2O</p> | |
| − | + | <p>32 g agar</p> | |
| − | + | <p>Autoclave</p> | |
| − | + | <p> </p> | |
| − | + | <p>After autoclaving the medium must cool down to ca. 50˚C. Now glycerol, IPTG, BCIP and the respective antiobiotic can be added.</p> | |
| − | + | <p> </p> | |
| − | + | <p>For 2 L medium:</p> | |
| − | + | <p>2 ml BCIP</p> | |
| − | + | <p>2 ml 1M IPTG</p> | |
| − | + | <p>2 ml Ampicllin or Kanamycin</p> | |
| − | + | <p>40 mL Glycerol</p> | |
| − | + | <p>The media is now ready for plating</p> | |
| − | + | <p><br /> <strong>Result</strong>: On Sperber medium phosphatase-recombinant colonies should develop a distinct color blue after 2 days.</p> | |
| − | + | <p> </p> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</div> | </div> | ||
| + | <a href="" onClick=" $('#menu14').slideToggle(300, function callback() { }); return false;"><h1>Esterase Activity plates, with 1% Tributyrin</h1></a> | ||
| + | <div id="menu14"> | ||
| + | <p style="text-align: center;"><span style="font-size: x-large; color: #888888;"><strong><span lang="EN-GB"></span></strong></span></p> | ||
| + | <p><span lang="EN-GB"> </span></p> | ||
| + | <p><span lang="EN-GB">To 500 ml of LB Media add 7.5 g of Agar and 5 ml of Tributyrin and homogenize with a mixer.</span></p> | ||
| + | <p><span lang="EN-GB">This culture medium must be directly sterilized by autoclaving at 121˚C for 20 min.</span></p> | ||
| + | <p><span lang="EN-GB">If you wait too long it will be inhomogeneous again!</span></p> | ||
| + | <p><span lang="EN-GB">When the medium cools down to 50°C, the antibiotic can be added.</span></p> | ||
| + | <p><span lang="EN-GB"> </span></p> | ||
| + | <p><strong><span lang="EN-GB">Result:</span></strong><span lang="EN-GB"> Halo formation is visible around the positive clones.</span></p> | ||
| + | </ul> | ||
| + | </div> | ||
| + | <a href="" onClick=" $('#menu12').slideToggle(300, function callback() { }); return false;"><h1> Cellulase activity plates</h1></a> | ||
| + | <div id="menu12"> | ||
| + | <p align="center"><span style="font-size: x-large; color: #828282;"><strong></strong></span></p> | ||
| + | <p><strong>A: LB_ agar base</strong></p> | ||
| + | <ul> | ||
| + | <li>Add the following components </li> | ||
| + | </ul> | ||
| + | <p> </p> | ||
| + | <table border="1" cellspacing="0" cellpadding="0"> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td valign="top"> | ||
| + | <p>Yeast extract </p> | ||
| + | </td> | ||
| + | <td valign="top"> | ||
| + | <p>5 g </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top"> | ||
| + | <p>Tryptone </p> | ||
| + | </td> | ||
| + | <td valign="top"> | ||
| + | <p>10 g </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top"> | ||
| + | <p>NaCl </p> | ||
| + | </td> | ||
| + | <td valign="top"> | ||
| + | <p>10 g </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top"> | ||
| + | <p>H<sub>2</sub>O </p> | ||
| + | </td> | ||
| + | <td valign="top"> | ||
| + | <p>Add to 1 L </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top"> | ||
| + | <p>Agar</p> | ||
| + | </td> | ||
| + | <td valign="top"> | ||
| + | <p>15 g</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | <p> </p> | ||
| + | <ul> | ||
| + | <li>Make sure to put a stirrer in the bottle.</li> | ||
| + | <li>Sterilize the culture medium by autoclaving at 121 <sup>o</sup>C for 20 min. </li> | ||
| + | </ul> | ||
| + | <p><strong> </strong></p> | ||
| + | <p><strong>B: Substrate for cellulase screening</strong></p> | ||
| + | <p>In 15 ml falcon tube, the substrate with final concentration 1% (w/v) is prepared in 96% (v/v) ethanol. For-example, to prepare 10 ml stock solution, weight 0.1 g of AZCL-HE-Cellulose and transfer it to 15 ml falcon tube, then add 96% (v/v) ethanol until the volume reached 10 ml.</p> | ||
| + | <p>The stock solution will be stored at +4 °C.</p> | ||
| + | <p><strong>C: Prepare the agar plates</strong></p> | ||
| + | <p>When the medium cools down to around 60 °C, put the bottle containing LB-agar base on a magnetic mixer. At the same time, disperse the substrate by flip the falcon tube several times. Then, pour the substrate into the LB-agar base gently until the substrate suspend evenly in the agar medium (For how much substrate should be poured, ask the supervisor). Other reagents like antibiotic can also be added. Ca. 3-5 ml substrate per Liter medium.</p> | ||
| + | <p><strong>D: Preparation of agar plates contacting cellulose substrate</strong></p> | ||
| + | <p>After dispersing substrate and adding antibiotic to LB-agar , pour the medium quickly with continuous stirring onto the plates and make sure to make a thin layer of medium. Let the plates to dry and store them at 4 °C.</p> | ||
| + | <p> </p> | ||
| + | <p><strong>E: Another method of pouring Substrate for cellulase screening on to the agar plates</strong></p> | ||
| + | <p>Rather than using normal pouring method one can first prepare regular LB plates having just a thin layer of LB. After preparation of substrate for cellulase screening add the proper antibiotic to it and then pour this mixture on top of this thin LB layer with constant stirring. Let the plates to dry and then store at 4 °C.</p> | ||
| + | <p><strong> </strong></p> | ||
| + | <p><strong> </strong></p> | ||
| + | <p><strong> </strong></p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | </div> | ||
| + | <a href="" onClick=" $('#menu33').slideToggle(300, function callback() { }); return false;"><h1 style="color:white;">1x TAE Buffer</h1></a> | ||
| + | <div id="menu33"> | ||
| + | <p>1 x TAE Buffer</p> | ||
| + | <table border="1" cellspacing="0" cellpadding="0"> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td valign="top" width="307"> | ||
| + | <p>Tris</p> | ||
| + | </td> | ||
| + | <td valign="top" width="307"> | ||
| + | <p>24.22 g</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="307"> | ||
| + | <p>Acetic acid (100%)</p> | ||
| + | </td> | ||
| + | <td valign="top" width="307"> | ||
| + | <p>5.74 ml</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="307"> | ||
| + | <p>EDTA (0.5M, pH 8.0)</p> | ||
| + | </td> | ||
| + | <td valign="top" width="307"> | ||
| + | <p>10 ml</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="307"> | ||
| + | <p>H<sub>2</sub>O dest.</p> | ||
| + | </td> | ||
| + | <td valign="top" width="307"> | ||
| + | <p>ad. 5 L</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | <p> </p> | ||
| + | </div> | ||
| − | + | <h2> Cloning Methods </h2> | |
| + | <a href="" onClick=" $('#menu6').slideToggle(300, function callback() { }); return false;"><h1>PCR product purification using QIAquick® PCR Purification Kit (QIAGEN)</h1></a> | ||
| + | <div id="menu6"> | ||
| − | |||
| − | |||
<ul> | <ul> | ||
| − | |||
<li>Add ethanol (96–100%) to Buffer PE before use.</li> | <li>Add ethanol (96–100%) to Buffer PE before use.</li> | ||
| − | <li>All centrifugation steps are carried out at | + | <li>All centrifugation steps are carried out at 17900 g in a conventional table-top microcentrifuge at room temperature.</li> |
</ul> | </ul> | ||
<p> </p> | <p> </p> | ||
| − | <p>- Add | + | <p>- Add 5 volumes Buffer PB to 1 volume of the PCR reaction and mix.</p> |
| − | + | <p>- Place a QIAquick column in a provided 2 ml collection tube.</p> | |
| − | + | <p>- To bind DNA, apply the sample to the QIAquick column and centrifuge for 30–60 s. Discard flow-through and place the QIAquick column back in the same tube.</p> | |
| − | + | <p>- To wash, add 0.75 ml Buffer PE to the QIAquick column, centrifuge for 30–60 s. Discard flow-through and place the QIAquick column back in the same tube.</p> | |
| − | <p>- | + | <p>- Centrifuge the QIAquick column once more in the provided 2 ml collection tube for 1 min to remove residual wash buffer.</p> |
| − | + | <p>- Place each QIAquick column in a clean 1.5 ml microcentrifuge tube.</p> | |
| − | + | <p>- To elute DNA, add 50 μl water (40 – 60 <sup>o</sup>C) to the center of the QIAquick membrane, let the column stand for 1 min, and then centrifuge for 1 min.</p> | |
| − | + | ||
| − | <p>- | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | <p>- | + | |
| − | + | ||
| − | <p>- | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | <p>- Place each QIAquick column in a clean 1.5 ml microcentrifuge tube.</p> | + | |
| − | + | ||
| − | <p>- To elute DNA, add 50 μl water (40 – 60 <sup>o</sup>C) to the center of the QIAquick membrane, let the column stand for 1 min, and then centrifuge for 1 min.</p> | + | |
| − | + | ||
</div> | </div> | ||
| − | + | <a href="" onClick=" $('#menu10').slideToggle(300, function callback() { }); return false;"><h1>PCR Gel extraction, peqGOLD Gel Extraction Kit</h1></a> | |
| − | + | <div id="menu10"> | |
| − | <a href="" onClick=" $('#menu4').slideToggle(300, function callback() { }); return false;"><h1>Blunt End Ligation in pJET1.2 vector –Clone JET PCR Cloning Kit–Thermo Scientific</h1></a> | + | <p> </p> |
| + | <p>- Fractionate DNA fragments by running a agarose gel (Do not stain the gel with Ethidium bromide or expose the DNA to UV for too long).</p> | ||
| + | <p>- Add equal volume of Binding Buffer to the gel slice and incubate at 65ᵒC for 8 min. Vortex or mix every 2 to 3 min until the agarose dissolves completely. (0.2 g of gel equivalent to 0.2 ml)</p> | ||
| + | <p>- Pour the mixture into PerfectBind DNA column (which is placed in a 2 ml collection tube) and centrifuge for 1 min at 10000 x g.(max. 750 µl) Discard the flow-through and place the PerfectBind DNA column in the same tube. Repeat the steps if required.</p> | ||
| + | <p>- Add 300 µl of binding Buffer to the PerfectBind DNA column for the washing the contaminants and centrifuge for 1 min at 10000 x g. Discard the flow-through and place the column in the same tube</p> | ||
| + | <p>- Add 750 µl of CG Wash Buffer to the PerfectBind DNA column for the wash, incubate for 2 to 3 min and centrifuge for 1 min at 10000 x g. Discard the flow-through and place the column in the same tube. Repeat this step once more.</p> | ||
| + | <p>- Centrifuge the PerfectBind DNA column once more in the 2 ml collection tube for 1 min at 10000 x g to remove the residual wash buffer.</p> | ||
| + | <p>- Place the PerfectBind DNA column in a clean 1.5 ml eppendorf tube.</p> | ||
| + | <p>- Add 50 µl of pre-warmed sterile water to the PerfectBind DNA column and incubate for 2 to 3 min (normally in a 2 step process of 30 µl in first elution step and 20 µl in second elution step) and centrifuge at 5000 x g for 1 min.</p> | ||
| + | <p>- Store the purified DNA at -20ᵒC.</p> | ||
| + | <p> </p> | ||
| + | <p></p> | ||
| + | </div> | ||
| + | <a href="" onClick=" $('#menu4').slideToggle(300, function callback() { }); return false;"><h1>Blunt End Ligation in pJET1.2 vector –Clone JET PCR Cloning Kit– (Thermo Scientific)</h1></a> | ||
<div id="menu4"> | <div id="menu4"> | ||
<p> </p> | <p> </p> | ||
| Line 211: | Line 448: | ||
</td> | </td> | ||
<td valign="top" width="246"> | <td valign="top" width="246"> | ||
| − | <p>10µl</p> | + | <p>10 µl</p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 219: | Line 456: | ||
</td> | </td> | ||
<td valign="top" width="246"> | <td valign="top" width="246"> | ||
| − | <p> | + | <p>125 ng</p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 227: | Line 464: | ||
</td> | </td> | ||
<td valign="top" width="246"> | <td valign="top" width="246"> | ||
| − | <p> | + | <p>50 ng (1µl)</p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 235: | Line 472: | ||
</td> | </td> | ||
<td valign="top" width="246"> | <td valign="top" width="246"> | ||
| − | <p>1µl</p> | + | <p>1 µl</p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 243: | Line 480: | ||
</td> | </td> | ||
<td valign="top" width="246"> | <td valign="top" width="246"> | ||
| − | <p>xµl</p> | + | <p>x µl</p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 251: | Line 488: | ||
</td> | </td> | ||
<td valign="top" width="246"> | <td valign="top" width="246"> | ||
| − | <p>20µl</p> | + | <p>20 µl</p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 260: | Line 497: | ||
<p>Cloning principle:</p> | <p>Cloning principle:</p> | ||
<ul> | <ul> | ||
| − | <li>pJET1.2 is a linearized cloning vector | + | <li>pJET1.2 is a linearized cloning vector designed for inserts from 6 bp to 10 kb. The 5’-ends of the vector contain phosporyl groups, therefore phosphorylation of the PCR products is indicated.</li> |
</ul> | </ul> | ||
<ul> | <ul> | ||
| Line 266: | Line 503: | ||
</ul> | </ul> | ||
<ul> | <ul> | ||
| − | <li>Optimal PCR product quantity for ligation reaction is to be calculated from the Kit protocol, For the length of | + | <li>Optimal PCR product quantity for ligation reaction is to be calculated from the Kit protocol, For the length of 2.5 kb of PCR product, to have 0.15 pmol ends of the PCR product in the ligation reaction 125 ng of the PCR product should be used.</li> |
</ul> | </ul> | ||
<ul> | <ul> | ||
| Line 273: | Line 510: | ||
</div> | </div> | ||
| − | <a href="" onClick=" $('#menu5').slideToggle(300, function callback() { }); return false;"><h1>TOPO® Cloning protocol usingChampion™ pET Directional TOPO® Expression Kits | + | <a href="" onClick=" $('#menu32').slideToggle(300, function callback() { }); return false;"><h1>Sticky End T4 Ligation (Thermo Scientific)</h1></a> |
| + | <div id="menu32"> | ||
| + | <p><strong>T4 Ligation - Thermo Fisher Scientific - sticky end ligation</strong></p> | ||
| + | <p>1. Prepare the following reaction mixture:</p> | ||
| + | <table border="1" cellspacing="0" cellpadding="0"> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td valign="top" width="184"> | ||
| + | <p>Linear vector DNA</p> | ||
| + | </td> | ||
| + | <td valign="top" width="104"> | ||
| + | <p align="center">20 ng</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="184"> | ||
| + | <p>Insert DNA</p> | ||
| + | </td> | ||
| + | <td valign="top" width="104"> | ||
| + | <p align="center">80 ng</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="184"> | ||
| + | <p>10x T4 DNA Ligase buffer</p> | ||
| + | </td> | ||
| + | <td valign="top" width="104"> | ||
| + | <p align="center">2 µl</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="184"> | ||
| + | <p>Thermo Scientific T4 DNA Ligase (Cat #EL0016)</p> | ||
| + | </td> | ||
| + | <td valign="top" width="104"> | ||
| + | <p align="center">1 U</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="184"> | ||
| + | <p>Nuclease free H<sub>2</sub>O</p> | ||
| + | </td> | ||
| + | <td valign="top" width="104"> | ||
| + | <p align="center">Add to 20 µl</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | <p>2. Incubate 1 hour at RT</p> | ||
| + | <p>3. Heat inactivation of T4 DNA Ligase at 65 °C for 10 minutes or at 70 °C for 5 min</p> | ||
| + | </div> | ||
| + | |||
| + | <a href="" onClick=" $('#menu5').slideToggle(300, function callback() { }); return false;"><h1>TOPO® Cloning protocol usingChampion™ pET Directional TOPO® Expression Kits (Thermo Fisher Scientific)</h1></a> | ||
<div id="menu5"> | <div id="menu5"> | ||
<ul> | <ul> | ||
| − | <li>You will perform TOPO® Cloning in a reaction buffer containing salt. Note | + | <li>You will perform TOPO® Cloning in a reaction buffer containing salt. Note: the amount of salt added to the TOPO® Cloning reaction has to be adapted to the planned transformation protocol (chemically competent cells or electrocompetent cells).</li> |
</ul> | </ul> | ||
<p><strong> </strong>- Set up the TOPO® Cloning reaction depending on the transformation method.</p> | <p><strong> </strong>- Set up the TOPO® Cloning reaction depending on the transformation method.</p> | ||
| Line 328: | Line 617: | ||
<tr> | <tr> | ||
<td valign="top" width="174"> | <td valign="top" width="174"> | ||
| − | <p>Sterile | + | <p>Sterile water</p> |
</td> | </td> | ||
<td valign="top" width="212"> | <td valign="top" width="212"> | ||
| Line 368: | Line 657: | ||
</ul> | </ul> | ||
</div> | </div> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<a href="" onClick=" $('#menu7').slideToggle(300, function callback() { }); return false;"><h1>Plasmid transformation into <strong>chemically competent <em>E. coli</em></strong></h1></a> | <a href="" onClick=" $('#menu7').slideToggle(300, function callback() { }); return false;"><h1>Plasmid transformation into <strong>chemically competent <em>E. coli</em></strong></h1></a> | ||
<div id="menu7"> | <div id="menu7"> | ||
| Line 407: | Line 678: | ||
<p> </p> | <p> </p> | ||
</div> | </div> | ||
| + | <a href="" onClick=" $('#menu11').slideToggle(300, function callback() { }); return false;"><h1>Electroporation of BL21 cells with pJET_RFP</h1></a> | ||
| + | <div id="menu11"> | ||
| + | <p> </p> | ||
| + | <p>- Apply microdialysis for 30 -45 min by applying the transformation mixture (plasmid+buffer) on (Millipore® MF-Millipore™ DNA Fillter Paper for Dialysis of DNA and Proteins – capitol scientific) after placing the membrane on sterile water.</p> | ||
| + | <p>- Defreeze 40 µl aliquots of chemically competent cells on ice</p> | ||
| + | <p>- Mix 50-200 ng DNA with the cells. (desalted)</p> | ||
| + | <p>- Transfer the culture without any air bubbles into pre-cooled electroporation cuvettes (40 µl maximum) and incubate on ice for 10 minutes.</p> | ||
| + | <p>- Electroporate using the electroporator with 1.25 mV, 5 decharge time.</p> | ||
| + | <p>- Immediately after electroporation, transfer 300 µl room temperature LB medium on top of the cells and transfer it into an 1.5 ml fresh E-cup</p> | ||
| + | <p>- Incubate the culture for 1 hour at 37°C and 150 rpm</p> | ||
| + | <p><sup>- </sup>Spread a 100 µl from the dilution series (10<sup>-3</sup> to 10<sup>-6</sup>) on a pre-warmed LB plate containing ampicillin</p> | ||
| + | <p><sup>- </sup>Incubate the plates overnight at 37°C </p> | ||
| + | <p> </p> | ||
| + | </div> | ||
| + | |||
| + | <a href="" onClick=" $('#menu3').slideToggle(300, function callback() { }); return false;"><h1>Plasmid Extraction - using QIAprep Spin Miniprep Kit (QIAGEN)</h1></a> | ||
| + | |||
| + | <div id="menu3"> | ||
| + | <p>This protocol describes the purification of plasmid DNA from 5 ml overnight cultures of <em>E. coli</em> grown in LB medium using the QIAprep Spin Miniprep Kit (QIAGEN)</p> | ||
| + | <ul> | ||
| + | <li>Add the provided RNase A solution to buffer P1, mix, and store at 4 <sup>o</sup>C.</li> | ||
| + | <li>Add ethanol (96–100%) to Buffer PE before use.</li> | ||
| + | <li>All centrifugation steps are carried out at 17,900 x g (13,000 rpm) in a conventional table-top microcentrifuge at room temperature.</li> | ||
| + | </ul> | ||
| + | <p> </p> | ||
| + | <p>- Add the proper antibiotic with the proper concentration to 5 ml LB medium (e.g Ampicilin is added to a final concentration of 100 µg/ml).</p> | ||
| + | |||
| + | <p>- Inoculate the medium with the desired <em>E.coli</em> strain and incubate overnight at 37 <sup>o</sup>C with agitation (150 rpm).</p> | ||
| + | |||
| + | <p>- Pellet the overnight culture by centrifugation at 8,000 rpm (6,800xg) for 3 min at room temperature.</p> | ||
| + | |||
| + | <p>- Resuspend pelleted bacterial cells in 250 μl Buffer P1 and transfer it to a microcentrifuge tube.</p> | ||
| + | |||
| + | <p>- Add 250 μl Buffer P2 and mix thoroughly by inverting the tube 4–6 times. Do not allow the lysis reaction to proceed for more than 5 min.</p> | ||
| + | |||
| + | <p>- Add 350 μl Buffer N3 and mix immediately and thoroughly by inverting the tube 4–6 times.</p> | ||
| + | |||
| + | <p>- Centrifuge for 10 min at 17,900 x g in a table-top microcentrifuge.</p> | ||
| + | |||
| + | <p>- Apply the supernatant to the QIAprep spin column by decanting. Centrifuge 60 s. Discard the flow-through.</p> | ||
| + | |||
| + | <p>- Wash the QIAprep spin column by adding 500 μl Buffer PB and centrifuging for 60 s. Discard the flow-through.</p> | ||
| + | |||
| + | <p>- Wash QIAprep spin column by adding 750 μl Buffer PE and centrifuging for 30–60 s.</p> | ||
| + | |||
| + | <p>- Discard the flow-through, and centrifuge for an additional 1 min to remove residual wash buffer.</p> | ||
| + | |||
| + | <p>- Place each QIAquick column in a clean 1.5 ml microcentrifuge tube.</p> | ||
| + | |||
| + | <p>- To elute DNA, add 50 μl water (40 – 60 <sup>o</sup>C) to the center of the QIAquick membrane, let the column stand for 1 min, and then centrifuge for 1 min.</p> | ||
| + | |||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | <a href="" onClick=" $('#menu2').slideToggle(300, function callback() { }); return false;"><h1>Plasmid Extraction - using peqGOLD Plasmid Miniprep Kit I (PEQLAB Technologies)</h1></a> | ||
| + | |||
| + | <div id="menu2"> | ||
| + | <p> | ||
| + | Select few TOP10/BL21 <em>E.coli</em> transformed colonies from the LB Amp plates containing the propagated transformed colonies and inoculate them into | ||
| + | liquid LB medium (5 ml) containing preferred antibiotic of desired concentration in each tube. | ||
| + | </p> | ||
| + | <p> | ||
| + | Incubate the LB tubes with the transformed colonies at 37°C in a shaker for about 12 – 15 hours (overnight) with agitation (150 rpm). | ||
| + | </p> | ||
| + | <p> | ||
| + | Extract the plasmids from the incubated LB cultures using the peqGOLD Plasmid Miniprep Kit I. | ||
| + | </p> | ||
| + | <p> | ||
| + | Centrifuge the culture at 10,000 x g for 2 min to obtain the pellet and repeat the process until the culture is completely centrifuged. Store the pellet of | ||
| + | 1 ml of the culture at -20°C for future use. | ||
| + | </p> | ||
| + | <p> | ||
| + | Resuspend the pellet in 250 µl of Solution I of the Kit (which is normally kept at 4°C because of the RNase) and vortex until the pellet is resuspended. | ||
| + | </p> | ||
| + | <p> | ||
| + | Add 250 µl of Solution II to the resuspended mixture and gently mix by inverting and rotating the tubes 6 -10 times to obtain a cleared lysate. Incubate | ||
| + | the mixture for 2 min to obtain optimum results. | ||
| + | </p> | ||
| + | <p> | ||
| + | Add 350 µl of Solution III to the cleared lysate and gently mix by inverting the tubes 6 -10 times until a flocculent white precipitate is formed. | ||
| + | Centrifuge at 10,000 x g for 10 min at room temperature. | ||
| + | </p> | ||
| + | <p> | ||
| + | Transfer the clear supernatant to a fresh PerfectBind DNA Column in a 2 ml Collection Tube. Centrifuge the Column with the Collection Tube for 1 min at | ||
| + | 10,000 x g at room temperature. Discard the flow-through liquid. | ||
| + | </p> | ||
| + | <p> | ||
| + | Add 500 µl of PW Plasmid buffer to the PerfectBind DNA Column in the Collection Tube and centrifuge for 1 min at 10,000 x g. Discard the flow-through. | ||
| + | </p> | ||
| + | <p> | ||
| + | Add 750 µl of DNA Wash buffer to the PerfectBind DNA Column in the Collection tube and centrifuge for 1 min at 10,000 x g. Discard the flow-through. Repeat | ||
| + | this step to obtain optimum results. | ||
| + | </p> | ||
| + | <p> | ||
| + | Place the PerfectBind DNA Column in the Collection tube and centrifuge for 2 min at 10,000 x g to dry the column matrix. This step is essential to remove | ||
| + | ethanol from the column. | ||
| + | </p> | ||
| + | <p> | ||
| + | Place the PerfectBind DNA Column into a fresh 1.5 ml Eppendorf tube. Add 50 µl of pre-warmed sterile deionized water directly to the binding matrix in the | ||
| + | PerfectBind DNA Column and centrifuge for 1 min at 5,000 x g to elute the DNA. | ||
| + | </p> | ||
| + | <p> | ||
| + | Discard the PerfectBind DNA Column and store the eluted plasmid DNA at -20°C. | ||
| + | </p> | ||
| + | <p> | ||
| + | Check the concentration of the plasmids by using a NanoDrop and note down the values for future experiments. | ||
| + | </p> | ||
| + | </div> | ||
| + | |||
| + | <h2> Competent Cells </h2> | ||
<a href="" onClick=" $('#menu8').slideToggle(300, function callback() { }); return false;"><h1>Preparation of competent <em>E.coli </em>cells</h1></a> | <a href="" onClick=" $('#menu8').slideToggle(300, function callback() { }); return false;"><h1>Preparation of competent <em>E.coli </em>cells</h1></a> | ||
<div id="menu8"> | <div id="menu8"> | ||
| Line 418: | Line 800: | ||
<p> pH 7.5 with acetic acid</p> | <p> pH 7.5 with acetic acid</p> | ||
<p>Buffer RF2 (80 ml): 1.6 ml MOPS (0.5 M) stock solution</p> | <p>Buffer RF2 (80 ml): 1.6 ml MOPS (0.5 M) stock solution</p> | ||
| − | <p> | + | <p> pH 6.8 with NaOH</p> |
<p> 0.096 g RbCl</p> | <p> 0.096 g RbCl</p> | ||
<p> 0.88 g CaCl<sub>2</sub>.2H<sub>2</sub>O</p> | <p> 0.88 g CaCl<sub>2</sub>.2H<sub>2</sub>O</p> | ||
| Line 424: | Line 806: | ||
<p>- Inoculate a 4 ml culture either with a single colony or with a cryoculture of the desired <em>E.coli </em>strainand incubate the culture with agitation overnight at 37°C</p> | <p>- Inoculate a 4 ml culture either with a single colony or with a cryoculture of the desired <em>E.coli </em>strainand incubate the culture with agitation overnight at 37°C</p> | ||
<p><sub>- </sub>Inoculate a 300 ml shake flask containing 100 ml LB medium with the overnight culture to an OD<sub>600</sub> of 0.05 and grow the culture at 37°C until the OD<sub>600</sub>is about 0.3</p> | <p><sub>- </sub>Inoculate a 300 ml shake flask containing 100 ml LB medium with the overnight culture to an OD<sub>600</sub> of 0.05 and grow the culture at 37°C until the OD<sub>600</sub>is about 0.3</p> | ||
| − | <p>- Transfer the cells into two 50 ml falcon tubes, incubate the cultures for 15 minues on ice and harvest the cells by centrifugation for 15 min at | + | <p>- Transfer the cells into two 50 ml falcon tubes, incubate the cultures for 15 minues on ice and harvest the cells by centrifugation for 15 min at 5,000 rpm and 4°C. Discard the supernatants.</p> |
| − | <p>- Resuspend the cells in 1/3 of the original volume (~16 ml/50 ml) of buffer RF1, incubate the cells again on ice and harvest the cells by centrifugation for 15 min at | + | <p>- Resuspend the cells in 1/3 of the original volume (~16 ml/50 ml) of buffer RF1, incubate the cells again on ice and harvest the cells by centrifugation for 15 min at 5,000 rpm and 4°C. Discard the supernatants.</p> |
<p>- Resuspend the cells in 4 ml of buffer RF2 and incubate the suspensions for 15 min on ice. Prepare the Eppendorf tubes and liquid nitrogen.</p> | <p>- Resuspend the cells in 4 ml of buffer RF2 and incubate the suspensions for 15 min on ice. Prepare the Eppendorf tubes and liquid nitrogen.</p> | ||
<p>- Put 0.4 ml of the cell suspension into the Eppendorf reaction tubes and freeze the cells by transferring them immediately to the liquid nitrogen. Store the competent cells at -80°C</p> | <p>- Put 0.4 ml of the cell suspension into the Eppendorf reaction tubes and freeze the cells by transferring them immediately to the liquid nitrogen. Store the competent cells at -80°C</p> | ||
| Line 451: | Line 833: | ||
<p>- Inoculate a 20 ml culture either with a single colony or with the cryoculture of the desired <em>E.coli </em>strainand incubate the culture with agitation for 20 h at 28°C</p> | <p>- Inoculate a 20 ml culture either with a single colony or with the cryoculture of the desired <em>E.coli </em>strainand incubate the culture with agitation for 20 h at 28°C</p> | ||
<p>- Inoculate a 250 ml SOB medium supplemented in a 2 l shake flask and grow the cells to an OD<sub>600</sub> of 0.5- 0.9 (20-20 h) at 18°C and 200-250 rpm</p> | <p>- Inoculate a 250 ml SOB medium supplemented in a 2 l shake flask and grow the cells to an OD<sub>600</sub> of 0.5- 0.9 (20-20 h) at 18°C and 200-250 rpm</p> | ||
| − | <p>- Incubate the whole flask for 10 min on ice. Collect the cells by centrifugation for 10 min at 4°C and 5000 rpm. Resuspend the cells in 80 ml of ice-cold TB and incubate them for 10 min on ice. Collect the cells by centrifugation for 5 min at | + | <p>- Incubate the whole flask for 10 min on ice. Collect the cells by centrifugation for 10 min at 4°C and 5000 rpm. Resuspend the cells in 80 ml of ice-cold TB and incubate them for 10 min on ice. Collect the cells by centrifugation for 5 min at 5,000 rpm.</p> |
<p>- Resuspend the cells in a 20 ml of ice-cold TB. Add DMSO to a final concentration of 7% (1.4 ml) and gently shake the falcon tube.</p> | <p>- Resuspend the cells in a 20 ml of ice-cold TB. Add DMSO to a final concentration of 7% (1.4 ml) and gently shake the falcon tube.</p> | ||
<p>- Transfer 0.2 ml aliquots into labelled eppendorf reaction tubes and freeze the cells in liquid nitrogen. Store the cells at –80°C.</p> | <p>- Transfer 0.2 ml aliquots into labelled eppendorf reaction tubes and freeze the cells in liquid nitrogen. Store the cells at –80°C.</p> | ||
| Line 457: | Line 839: | ||
<p> </p> | <p> </p> | ||
</div> | </div> | ||
| − | <a href="" onClick=" $('# | + | <a href="" onClick=" $('#menu16').slideToggle(300, function callback() { }); return false;"> |
| + | <h1>Transformation Efficiency Kit, RFP construct (iGEM)</h1></a> | ||
| + | <div id="menu16"> | ||
| + | <p>Before using our competent <em>E. coli</em> TOP10 cells in the important experiments, we used the Transformation Efficiency Kit to test the efficiency of our competent cells!</p> | ||
| + | <p>The kit includes five vials of each different DNA concentration: 50pg/μl, 20pg/μl, 10pg/μl, 5pg/μl, 0.5pg/μl of purified DNA from BBa_J04450 (RFP construct) in plasmid backbone pSB1C3.</p> | ||
| + | <p> </p> | ||
| + | <p><strong>Protocol as distributed by iGEM (modified) </strong></p> | ||
| + | <p>Spin down the DNA tubes from the Transformation Efficiency Kit to collect all of the DNA into the bottom of each tube prior to use. A quick spin of 20-30 seconds at 8000-10000 rpm will be sufficient. Note: There should be 50 µL of DNA in each tube sent in the Kit.</p> | ||
| + | <p>Thaw competent cells on ice. Label one 2.0 ml microcentrifuge tube for each concentration and then pre-chill by placing the tubes on ice.</p> | ||
| + | <p>Pipet 1 µL of DNA into each microcentrifuge tube. For each concentration, use a separate tube.</p> | ||
| + | <p>Pipet 50 µL of competent cells into each tube. Flick the tube gently with your finger to mix. Incubate on ice for 30 minutes. Pre-heat heating block now to 42°C.</p> | ||
| + | <p>Heat-shock the cells by placing onto the heating block for 1 minute.</p> | ||
| + | <p>Immediately transfer the tubes back to ice, and incubate on ice for 5 minutes. This helps the cells recover.</p> | ||
| + | <p>Add 200 µL of LB media per tube, and incubate at 37°C for 1 hour. Prepare the LB-Cam (Chloramphenicol) agar plates during this time: label them.</p> | ||
| + | <p>Pipet 20 µL from each tube onto the appropriate plate, and spread the mixture evenly across the plate. Do triplicates (3 each) of each tube if possible, so you can calculate an average colony yield. Incubate at 37°C overnight. Position the plates so the agar side is facing up, and the lid is facing down.</p> | ||
| + | <p>Count the number of colonies on a light field or a dark background, such as a lab bench. Use the following equation to calculate your competent cell efficiency. If you've done triplicates of each sample, use the average cell colony count in the calculation.</p> | ||
| + | <p>(colonies on plate) / ng of DNA plated x 1,000ng/µg</p> | ||
| + | <p>Note: The measurement "ng of DNA plated" refers to how much DNA was plated onto each agar plate, not the total amount of DNA used per transformation. You can calculate this number using the following equation:</p> | ||
| + | <p>1 µL x concentration of DNA (refer to vial) x (volume plated / total reaction volume)</p> | ||
| + | <p><strong><p> </p>NOTE: Since this protocol led to a very low transformation efficiency we repeated the experiment with 200 µL competent cells instead of 50 µL and plated 100 µL instead of 20 µL.</strong></p> | ||
| + | <p><strong> </strong></p> | ||
| + | <p><strong>Results</strong></p> | ||
| + | <p>Competent cells should have an efficiency of 1.5x10<sup>8</sup> to 6x10<sup>8</sup> cfu/µg DNA, where "cfu" means "colony-forming unit" and is a measurement of cells.</p> | ||
| + | </div> | ||
| + | |||
| + | <h2> Protein Extraction and Purification</h2> | ||
| + | <a href="" onClick=" $('#menu36').slideToggle(300, function callback() { }); return false;"><h1 style="color:white;"> Induction, harvest and disruption of expression cultures</h1></a> | ||
| + | <div id="menu36"> | ||
| + | <p> | ||
| + | <u>Induction</u> | ||
| + | </p> | ||
| + | <p> | ||
| + | Recombinant protein expression was exclusively realized in <em>E.coli</em>. pET101-ScaA was expressed in BL21(DE3) cells under the control of the <em>lac</em> operon, and pBAD-RFP-ACEL (and all other pBAD constructs) was expressed in TOP 10 cells under the control of the <em>ara</em> operon. The | ||
| + | procedure is as follows:<u></u> | ||
| + | </p> | ||
| + | <p> | ||
| + | Inoculate “fat” LB expression medium, having a volume corresponding to 5 % of the shake flask that is used, with 5 % (v/v) of an overnight LB starter | ||
| + | culture grown at 37°C and 150 rpm. | ||
| + | </p> | ||
| + | <p> | ||
| + | Incubate the expression culture at 37°C and 150 rpm until an OD<sub>600</sub> between 2.5 and 3.5 is reached. | ||
| + | </p> | ||
| + | <p> | ||
| + | Induce protein expression by adding the appropriate inducer to the expression culture. In case of pET101-ScaA use 1 mM IPTG and in case of pBAD-RFP-ACEL | ||
| + | use 0.2 % (v/v) L-arabinose. | ||
| + | </p> | ||
| + | <p> | ||
| + | Incubate the expression culture for 20 h at 37°C and 150 rpm. | ||
| + | </p> | ||
| + | <p> | ||
| + | Take a 1 ml sample before induction and before cell harvesting in order to analyze the expression effectivity of target proteins by SDS-PAGE. | ||
| + | </p> | ||
| + | <p> | ||
| + | <u>Cell Harvest</u> | ||
| + | </p> | ||
| + | <p> | ||
| + | Pellet the culture at 4°C, 13000rpm for 20min (SLA-3000 rotor, Sorvall). | ||
| + | </p> | ||
| + | <p> | ||
| + | Resuspend and wash with an equal volume of LEW Buffer (see Protein extraction). | ||
| + | </p> | ||
| + | <p> | ||
| + | Repeat previous centrifugation step. | ||
| + | </p> | ||
| + | <p> | ||
| + | Freeze pellet until further use or prepare for French Press. | ||
| + | </p> | ||
| + | <p> | ||
| + | <u>Cell extraction by French Press</u> | ||
| + | </p> | ||
| + | <p> | ||
| + | Resuspend pellet in 20mL 1x LEW Buffer . | ||
| + | </p> | ||
| + | <p> | ||
| + | Take a small sample (10µl) for microscopy. | ||
| + | </p> | ||
| + | <p> | ||
| + | Disrupt cells by using a discontinuous high-pressure homogenizer (e.g., French-Press, 4 cycles at 1000 psi). | ||
| + | </p> | ||
| + | <p> | ||
| + | take a small sample (10µl) and analyse both before and after press samples under the microscope. Look for inclusion bodies. | ||
| + | </p> | ||
| + | <p> | ||
| + | Sediment cell debris via centrifugation for 30 min at 13000 rpm (SS-34 rotor, Sorvall) and 4°C. | ||
| + | </p> | ||
| + | <p> | ||
| + | Filter the resulting supernatant through 0.45 µm sterile filters and subsequently apply it <a name="_GoBack"></a>to chromatographic purification steps. | ||
| + | </p> | ||
| + | </div> | ||
| + | |||
| + | <a href="" onClick=" $('#menu29').slideToggle(300, function callback() { }); return false;"><h1 style="color:white;">Protein Purification (Protino® Ni-IDA 2000 His-Tag protein purification, Macherey-Nagel)</h1></a> | ||
| + | <div id="menu29"> | ||
| + | <ul> | ||
| + | <p></p> | ||
| + | <p>1 x LEW (Lysis-Equilibrium-Wash) Buffer:</p> | ||
| + | </ul> | ||
| + | <table border="1" cellspacing="0" cellpadding="0"> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td valign="top" width="307"> | ||
| + | <p>NaH<sub>2</sub>PO<sub>4</sub> x 2 H<sub>2</sub>O (MW: 156.07 g/mol)</p> | ||
| + | </td> | ||
| + | <td valign="top" width="97"> | ||
| + | <p>7.8 g</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="307"> | ||
| + | <p>NaCl (MW: 58.44 g/mol)</p> | ||
| + | </td> | ||
| + | <td valign="top" width="97"> | ||
| + | <p>17.5 g</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="307"> | ||
| + | <p>H<sub>2</sub>0 dest.</p> | ||
| + | </td> | ||
| + | <td valign="top" width="97"> | ||
| + | <p>ad. 1000 mL</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | <p> </p> | ||
| + | <p>Adjust to pH 8.0 using NaOH.</p> | ||
| + | <p>1 x Elution Buffer:</p> | ||
| + | <table border="1" cellspacing="0" cellpadding="0"> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td valign="top" width="307"> | ||
| + | <p>NaH<sub>2</sub>PO<sub>4</sub> x 2 H<sub>2</sub>O (MW: 156.07 g/mol)</p> | ||
| + | </td> | ||
| + | <td valign="top" width="97"> | ||
| + | <p>7.8 g</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="307"> | ||
| + | <p>NaCl (MW: 58.44 g/mol)</p> | ||
| + | </td> | ||
| + | <td valign="top" width="97"> | ||
| + | <p>17.5 g</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="307"> | ||
| + | <p>Imidazole (MW: 68.08 g/mol)</p> | ||
| + | </td> | ||
| + | <td valign="top" width="97"> | ||
| + | <p>17.0 g</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="307"> | ||
| + | <p>H<sub>2</sub>0 dest.</p> | ||
| + | </td> | ||
| + | <td valign="top" width="97"> | ||
| + | <p>ad. 1,000 mL</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | <p> </p> | ||
| + | <p>Adjust to pH 8.0 using NaOH.</p> | ||
| + | <p> </p> | ||
| + | |||
| + | <p><strong>Protino® Ni-IDA 2,000 His-Tag protein purification (Macherey-Nagel)</strong></p> | ||
| + | <p>- Wet Ni-IDA column with 4 mL of 1x LEW buffer and discard flow through</p> | ||
| + | <p>- Run supernatant from the cell extraction through the column and collect flow through (cell extract, store at 4°C)</p> | ||
| + | <p>- Wash column 3 times with 4 mL 1x LEW Buffer</p> | ||
| + | <p>- Collect each flow through (wash 1-3) and store at 4°C</p> | ||
| + | <p>- Elute protein 3 times with 3 mL Elution Buffer (contains Imidazole)</p> | ||
| + | <p>- Collect each elution and store fractions at 4°C</p> | ||
| + | <p>- Quantify protein content by Bradford measurement (see Bradford assay)</p> | ||
| + | </div> | ||
| + | |||
| + | <a href="" onClick=" $('#menu34').slideToggle(300, function callback() { }); return false;"><h1 style="color:white;"> Affinity chromatography of His-tagged proteins</h1></a> | ||
| + | <div id="menu34"> | ||
| + | <p> | ||
| + | The N-terminal His<sub>6</sub>- tagged fusion proteins His<sub>6</sub>-ScaA and His<sub>6</sub>-RFP-ACEL were purified by nickel affinity chromatography | ||
| + | using Ni-NTA-agarose columns in a first purification step. | ||
| + | </p> | ||
| + | <p> | ||
| + | <u>Preparation:</u> | ||
| + | </p> | ||
| + | <p> | ||
| + | Connect a 5 ml HisTrap FF column (GE Healthcare) to an Äkta purifier or Äkta prime system and wash the column with 1 CV (column volume) 20 % EtOH and a | ||
| + | maximum flow rate of 3 ml/min. | ||
| + | </p> | ||
| + | <p> | ||
| + | Pre-equilibrate the column with lysis buffer by first washing 3 CV with water, followed by 3 CV with buffer, each with a maximum flow rate of 5 ml/min. | ||
| + | </p> | ||
| + | <p> | ||
| + | <u>Run:</u> | ||
| + | </p> | ||
| + | <p> | ||
| + | 1.) Load the protein solution onto the column using an appropriate super loop (10 ml or 50 ml) and a maximum flow rate of 1 ml/min. Start the automated | ||
| + | fractionation system. | ||
| + | </p> | ||
| + | <p> | ||
| + | 2.) Once the protein is loaded, wash the column with lysis buffer using a maximum flow rate of 5 ml/min until the UV absorption has reached a constant | ||
| + | value. | ||
| + | </p> | ||
| + | <p> | ||
| + | 3.) Start eluting unspecifically bound, contaminative protein species with a flow rate of 3 ml/min by applying a stepwise gradient from 0 % to 2 % of | ||
| + | elution buffer. | ||
| + | </p> | ||
| + | <p> | ||
| + | 4.) Elute the target protein with a flow rate of 3 ml/min by applying a linear gradient from 2 % to 50 % of elution buffer over 20 CV (column volumes). | ||
| + | </p> | ||
| + | <p> | ||
| + | 5.) Analyze obtained peak fractions by SDS-PAGE and pool the fractions containing the target protein. | ||
| + | </p> | ||
| + | <p> | ||
| + | 6.) Reduce the volume of the protein solution via concentration until a maximum volume of 5 ml is reached and apply it to size exclusion chromatography. | ||
| + | </p> | ||
| + | <p> | ||
| + | All chromatographical purification steps are carried out at 20°C. | ||
| + | </p> | ||
| + | <p> | ||
| + | Required buffers | ||
| + | </p> | ||
| + | <p> | ||
| + | Buffer A: Lysis buffer (20 mM phosphate pH 8, 500 mM NaCl, 5 % (v/v) glycerol, 1 mM 2-mercaptoethanol) | ||
| + | </p> | ||
| + | <p> | ||
| + | Buffer B: Elution buffer (Lysis buffer supplemented with 250 mM imidazole) | ||
| + | </p> | ||
| + | |||
| + | </div> | ||
| + | <a href="" onClick=" $('#menu39').slideToggle(300, function callback() { }); return false;"><h1 style="color:white;"> Size Exclusion Chromatography</h1></a> | ||
| + | <div id="menu39"> | ||
| + | <p> | ||
| + | In a second purification step, the target proteins ScaA and RFP-ACEL were separated according to their size (radius of gyration) by employing size | ||
| + | exclusion chromatography. Therefore: | ||
| + | </p> | ||
| + | <p> | ||
| + | Connect a HiLoad Superdex 200 16/600 column (GE Healthcare) to an Äkta purifier or Äkta prime system and wash the column with 1 CV of 20 % EtOH and a | ||
| + | maximum flow rate of 1 ml/min. | ||
| + | </p> | ||
| + | <p> | ||
| + | Pre-equilibrate the column with loading buffer by washing first with 1 CV of water and subsequently with 1 CV of the respective buffer, each with a maximum | ||
| + | flow rate of 1 ml/min. | ||
| + | </p> | ||
| + | <p> | ||
| + | Load the concentrated protein solution with a maximum flow rate of 1 ml/min onto the column by using a 5 ml sampling loop. Start the automated | ||
| + | fractionation. | ||
| + | </p> | ||
| + | <p> | ||
| + | Elute the target protein with a maximum flow rate of 1 ml/min and analyze obtained peak fractions on a SDS gel. | ||
| + | </p> | ||
| + | <p> | ||
| + | Pool the fractions containing the target protein. | ||
| + | </p> | ||
| + | <p> | ||
| + | All chromatographic purification steps are carried out at room temperature. | ||
| + | </p> | ||
| + | <p> | ||
| + | Required buffer: | ||
| + | </p> | ||
| + | <p> | ||
| + | Buffer A: Loading buffer (20 mM phosphate pH 8, 500 mM NaCl, 5 % glycerol, 1 mM 2-mercaptoethanol) | ||
| + | </p> | ||
| + | |||
| + | </div> | ||
| + | <a href="" onClick=" $('#menu35').slideToggle(300, function callback() { }); return false;"><h1 style="color:white;"> Concentration of protein solutions</h1></a> | ||
| + | <div id="menu35"> | ||
| + | <p> | ||
| + | Concentration of protein solutions was realized via centrifugation using 20 ml Vivaspin centrifugal concentrators (Viva Science) with the appropriate | ||
| + | molecular weight cutoffs of 50 kDa in case of the scaffolding protein ScaA and 30 kDa in case of RFP-ACEL. In order to concentrate protein solutions: | ||
| + | </p> | ||
| + | <p> | ||
| + | Wash the concentrator with 20 ml of buffer in order to remove the protectant (glycerol) from the membrane. | ||
| + | </p> | ||
| + | <p> | ||
| + | Add the protein sample to the concentrator and start centrifugation at 4°C until the desired volume/concentration is reached. The sample volume and the | ||
| + | speed is determined by the rotor that is used. If a 25° fixed angle rotor is applied, the maximum starting volume is 14 ml and the speed is restricted to | ||
| + | 6000 x g. | ||
| + | </p> | ||
| + | <p> | ||
| + | Gently mix the upper reservoir prior to each refilling step and prior to transfer. | ||
| + | </p> | ||
| + | <p> | ||
| + | Check the flow through for leakage by Bradford assay. | ||
| + | </p> | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <a href="" onClick=" $('#menu30').slideToggle(300, function callback() { }); return false;"><h1 style="color:white;">Bradford Assay</h1></a> | ||
| + | <div id="menu30"> | ||
| + | <p><strong>Bradford Assay</strong></p> | ||
| + | <p>For calculating protein concentrations of an unknown sample the traditional way is to use a standard curve that is generated from known protein standards. Readily available protein standards contain bovine serum albumin (BSA) or bovine gamma globulin (γ-globulin, IgG). The calculated result is an estimation of protein concentration since Bradford protein assays do show significant protein-to-protein variation.</p> | ||
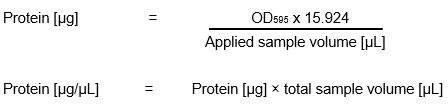
| + | <p>For our Bradford assay we got an already prepared standard solution with a factor of 15.924.</p><p>Caution: Store the Bradford solution in the dark, it is light sensitive.</p> | ||
| + | <p> </p> | ||
| + | <p><strong>Sample preparation:</strong></p> | ||
| + | <p>Measurement takes place against 20 μL water (blank).</p> | ||
| + | <p>Blank: 20 μL + 1 mL Bradford solution</p> | ||
| + | <p>Samples: 1-20 μL (depending on protein concentration, max. 2 mg/mL) + 19-0 μL water + 1 mL Bradford solution</p> | ||
| + | <ul> | ||
| + | <li>Prepare photometer cuvettes for technical triplicates of all samples + one blank</li> | ||
| + | <li>Preheat photometer light and set it to 595 nm</li> | ||
| + | <li>Pipet the samples (plus water) and the blank into the cuvettes</li> | ||
| + | <li>Pipet 1 mL Bradford solution on top, carefully homogenize by pipetting up and down</li> | ||
| + | <li>Avoid air bubbles</li> | ||
| + | <li>Incubate the cuvettes in the dark for 5 minutes</li> | ||
| + | <li>Immediately measure the samples at 595 nm</li> | ||
| + | </ul> | ||
| + | <p> </p> | ||
| + | <p>Calculation of protein concentration:</p> | ||
| + | </html> | ||
| + | |||
| + | [[File:Bradford_calculation_iGEM_Goettingen_2015.jpeg|thumb|center|300px|<p style="text-align: justify;">Formula for calculating the protein concentration</p>]] | ||
| + | |||
| + | <html> | ||
| + | <p><br /> </p> | ||
| + | <p> </p> | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <a href="" onClick=" $('#menu38').slideToggle(300, function callback() { }); return false;"><h1 style="color:white;"> SDS Polyacrylamid Gel Electrophoresis</h1></a> | ||
| + | <div id="menu38"> | ||
| + | <p> | ||
| + | The analytic protein separation according to the apparent molecular weight was carried out by discontinuous SDS polyacrylamid gel electrophoresis according | ||
| + | to Laemmli (1970) using mini gel electrophoresis chambers (BioRad, 10x8 cm). For cast and run of the gels do the following: | ||
| + | </p> | ||
| + | <p> | ||
| + | For the stacking gel use the following components: 0.25 % of the total volume of stacking gel buffer, and acrylamid to a final concentration of 4 %. Add | ||
| + | water to the final volume (cf. pipetting scheme). Add 1/133 of the total volume of 10 % ammonium persulfate (APS) and 1/1000 of | ||
| + | N,N,N’,N’-tetramethyl-ethylenediamine (TEMED). | ||
| + | </p> | ||
| + | <p> | ||
| + | For the separation gel mix the appropriate amount of acrylamid (e.g. 10 % for a 10 % SDS gel) with 0.25 % of the final volume of separation gel buffer and | ||
| + | fill with water to the final volume. Add 1/133 of the total volume of 10 % APS and 1/1000 of TEMED. | ||
| + | </p> | ||
| + | <p> | ||
| + | Place the poured gel into the running chamber and fill the upper and lower reservoir with running buffer. | ||
| + | </p> | ||
| + | <p> | ||
| + | Dilute the protein samples 1:1 with loading buffer and load the gel. | ||
| + | </p> | ||
| + | <p> | ||
| + | Run the gel with 15mA per gel for 15 min, and then increase to 30mA per Gel until the end. | ||
| + | </p> | ||
| + | <p> | ||
| + | After electrophoretic separation, place the gels into staining solution and heat it without boiling. | ||
| + | </p> | ||
| + | <p> | ||
| + | Incubate the gels under slow agitation until protein bands become visible. | ||
| + | </p> | ||
| + | <p> | ||
| + | Discolor the gels by using fixation solution. | ||
| + | </p> | ||
| + | <p> | ||
| + | <strong>Pipetting scheme. </strong> | ||
| + | </p> | ||
| + | <table border="1" cellspacing="0" cellpadding="0"> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | <strong>Compoment</strong> | ||
| + | </p> | ||
| + | <p align="center"> | ||
| + | <strong>(For 2 gels)</strong> | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | <strong>Stacking gel</strong> | ||
| + | </p> | ||
| + | <p align="center"> | ||
| + | <strong>(4 % acrylamid)</strong> | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | <strong>Separation gel</strong> | ||
| + | </p> | ||
| + | <p align="center"> | ||
| + | <strong>(10 % acrylamid)</strong> | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | Water | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | 2.64 ml | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | 4 ml | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | Acrylamid | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | 0.4 ml | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | 2 ml | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | Stacking gel buffer | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | 0.96 ml | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | --- | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | Separation gel buffer | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | --- | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | 2 ml | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="636" colspan="3" valign="top"> | ||
| + | <p align="center"> | ||
| + | <strong>Mix</strong> | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | 10 % APS | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | 30 µl | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | 60 µl | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | TEMED | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | 4 µl | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | 6 µl | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | <p> | ||
| + | Required agents and solutions: | ||
| + | </p> | ||
| + | <p> | ||
| + | 40 % Acrylamid/bis-acrylamid (37.5:1) | ||
| + | </p> | ||
| + | <p> | ||
| + | 10 % (w/v) SDS solution | ||
| + | </p> | ||
| + | <p> | ||
| + | Stacking gel buffer (1.5 M Tris/HCl pH 8.8, 0.4 % (w/v) SDS) | ||
| + | </p> | ||
| + | <p> | ||
| + | Separation gel buffer (0.5 M Tris/HCl pH 6.5, 0.4 % (w/v) SDS) | ||
| + | </p> | ||
| + | <p> | ||
| + | 2x Laemmli loading buffer (20 mM Tris/HCl pH 6.5, 4 % (w/v) SDS, 10 % (v/v) β-mercaptoethanol, 40 % (v/v) glycerol, 0.002 % (w/v) bromphenol blue) | ||
| + | </p> | ||
| + | <p> | ||
| + | 1x SDS running buffer (25 mM Tris base, 192 mM glycine, 0.1 % (w/v) SDS) | ||
| + | </p> | ||
| + | <p> | ||
| + | Staining solution (0.005 % (w/v) Coomassie Brilliant Blue G-250, 0.0025 % (w/v) Coomassie Brilliant Blue R-250, 10 % (v/v) ethanol und 5 % (v/v) acetic | ||
| + | acid) | ||
| + | </p> | ||
| + | <p> | ||
| + | Fixation solution (10 % (v/v) acetic acid) | ||
| + | </p> | ||
| + | </div> | ||
| + | |||
| + | <a href="" onClick=" $('#menu40').slideToggle(300, function callback() { }); return false;"><h1 style="color:white;"> Western Blot</h1></a> | ||
| + | <div id="menu40"> | ||
| + | <p> | ||
| + | 10 x TBS Buffer (store at RT) | ||
| + | </p> | ||
| + | <table border="1" cellspacing="0" cellpadding="0"> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | Tris HCL (MW: 121,14 g/mol) | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | 6.057g | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | NaCl (MW: 58.44 g/mol) | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | 40.91g | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | H<sub>2</sub>O dest. | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | ad. 500 ml | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | <p> | ||
| + | adjust to pH 7.4 | ||
| + | </p> | ||
| + | <p> | ||
| + | 1 x Transfer Buffer (store at RT) | ||
| + | </p> | ||
| + | <table border="1" cellspacing="0" cellpadding="0"> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | Tris | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | 3.029 g | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | Glycin | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | 14,41g | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | Methanol | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | 200ml | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | H<sub>2</sub>O bidest. | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | ad. 1000ml | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | <p> | ||
| + | check pH: 8.1-8.4 | ||
| + | </p> | ||
| + | <p> | ||
| + | If the pH is not correct, the Buffer has to be remade. Do not adjust. | ||
| + | </p> | ||
| + | <p> | ||
| + | 10 x TBS-Tween Buffer (store at RT) | ||
| + | </p> | ||
| + | <table border="1" cellspacing="0" cellpadding="0"> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | Tris-HCL (MW: 121.14 g/mol) | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | 6.057g | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | NaCl (MW: 58.44 g/mol) | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | 40.91g | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | Tween20 (Endkonz. 0,1%) | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | 0.5ml | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | H<sub>2</sub>O bidest. | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | ad. 500ml | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | <p> | ||
| + | adjust to pH 7.4 | ||
| + | </p> | ||
| + | <p> | ||
| + | Blocking Solution | ||
| + | </p> | ||
| + | <table border="1" cellspacing="0" cellpadding="0"> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | Skim Milk | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | 2,5g | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | 1 x TBS | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | 50ml | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | <p> | ||
| + | Dissolve for 15-30 min at RE. Store 400µl at 4°C for a later step. | ||
| + | </p> | ||
| + | <p> | ||
| + | Phosphatase Buffer (store at RT) | ||
| + | </p> | ||
| + | <table border="1" cellspacing="0" cellpadding="0"> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | Tris-HCL (MW: 121.14 g/mol) | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | 6.057g | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | NaCl (MW: 58.44 g/mol) | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | 2.922 g | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | MgCl<sub>2 </sub>x 6 H<sub>2</sub>O (MW: 203.3 g/mol) | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | 0.51 g | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | H<sub>2</sub>O bidest. | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | ad. 500ml | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | <p> | ||
| + | adjust to pH 9.5 | ||
| + | </p> | ||
| + | <p> | ||
| + | Antibody: Anti V5-AP Antibody (Invitrogen, store at 4°C) | ||
| + | </p> | ||
| + | <p> | ||
| + | NBT: Promega, 50 mg/ml, store at -20°C | ||
| + | </p> | ||
| + | <p> | ||
| + | BCIP: Promega, 50 mg/ml, store at -20°C | ||
| + | </p> | ||
| + | <p> | ||
| + | 1.) Run an SDS Gel and use Protein Ladder compatible with Western Blots. | ||
| + | </p> | ||
| + | <p> | ||
| + | 2.) Use Perfect Blue Semi-Dry Electro blotter from Peqlab and assemble apparatus in a tip box with a little transfer buffer (sandwich should be moist <u>not </u>wet): | ||
| + | </p> | ||
| + | <p> | ||
| + | Base of blotter = Kathode (+) bottom | ||
| + | </p> | ||
| + | <p> | ||
| + | 3x Whatman paper (moist) size of SDS page | ||
| + | </p> | ||
| + | <p> | ||
| + | Blotting membrane (Hybond EHL, 0,45µm, moist, no bubbles, SDS page sized) | ||
| + | </p> | ||
| + | <p> | ||
| + | SDS Gel (no bubbles! Proteins have a neg. charge and go towards the Kathode) | ||
| + | </p> | ||
| + | <p> | ||
| + | 3x Whatman paper (moist, gel sized) | ||
| + | </p> | ||
| + | <p> | ||
| + | Lid = Anode (-) top | ||
| + | </p> | ||
| + | <p> | ||
| + | After assembly of the sandwich the lid is screwed on lightly with 3 screws. Connect Blotter to an adequate power supply, which can work at a constant 10V. | ||
| + | Blot at 200mA per Gel and constant 10V for 1 hour. | ||
| + | </p> | ||
| + | <p> | ||
| + | 3.) Wash the membrane 3x 5 min with 1x TBS Buffer on a rocker | ||
| + | </p> | ||
| + | <p> | ||
| + | 4.) Block membrane in 50ml 1 x TBS Buffer with 5% skim milk for one hour or overnight on a rocker. | ||
| + | </p> | ||
| + | <p> | ||
| + | 5.) Wash 3x 15min 1xTBS-Tween on a rocker | ||
| + | </p> | ||
| + | <p> | ||
| + | 6.) Prepare Antibody: 14ml 1xTBS-Tween + 200µl 1x TBS Puffer/5% Skimmilk + 7µl Antibody | ||
| + | </p> | ||
| + | <p> | ||
| + | 7.) Incubate with antibody for 2 hours on a rocker | ||
| + | </p> | ||
| + | <p> | ||
| + | 8.) Wash 2x shortly in 1x TBS-Tween Buffer on a rocker | ||
| + | </p> | ||
| + | <p> | ||
| + | 9.) 1x 15min 1x TBS-Tween Buffer on the rocker | ||
| + | </p> | ||
| + | <p> | ||
| + | 10.) 2x 5min 1x TBS-Tween Buffer on the rocker | ||
| + | </p> | ||
| + | <p> | ||
| + | 11.) Wash 2x 10 min 1x TBS Buffer on a rocker | ||
| + | </p> | ||
| + | <p> | ||
| + | 12.) Wash 2x shortly in 1x Phosphatse buffer | ||
| + | </p> | ||
| + | <p> | ||
| + | 13.) 10ml 1x Phosphatase buffer + 66µl NBT + 33µl BCIP | ||
| + | </p> | ||
| + | <p> | ||
| + | - stain in the dark until bands become visible | ||
| + | </p> | ||
| + | <p> | ||
| + | - stop the reaction by washing with H2O | ||
| + | </p> | ||
| + | <p> | ||
| + | - dry western blot in Whatman Paper | ||
| + | </p> | ||
| + | <p> | ||
| + | 14.) Scan the membrane | ||
| + | </p> | ||
| + | |||
| + | </div> | ||
| + | <h2> Activity Screens & Tests</h2> | ||
| + | <a href="" onClick=" $('#menu9').slideToggle(400, function callback() { }); return false;"><h1>Esterase activity test</h1></a> | ||
<div id="menu9"> | <div id="menu9"> | ||
<p>Materials:</p> | <p>Materials:</p> | ||
| − | <p>Substrate- 1mM 4-nitrophenyl butyrate prepared in Na-Phosphate buffer ( | + | <p>Substrate- 1mM 4-nitrophenyl butyrate prepared in Na-Phosphate buffer (pH 8.0)</p> |
<p>Protein sample</p> | <p>Protein sample</p> | ||
<p>Procedure:</p> | <p>Procedure:</p> | ||
| Line 470: | Line 1,721: | ||
<p> </p> | <p> </p> | ||
</div> | </div> | ||
| − | + | ||
| − | <div id=" | + | <a href="" onClick=" $('#menu31').slideToggle(400, function callback() { }); return false;"><h1>Phosphatase activity test</h1></a> |
| − | + | <div id="menu31"> | |
| − | <p> | + | |
| − | < | + | <p><strong>Materials:</strong></p> |
| − | + | <ul> | |
| − | <p>- | + | <li>Freshly prepared AAM Solution (Acetone, 5N H<sub>2</sub>SO<sub>4</sub>, 10 mM Ammonium molybdate, 2:1:1 v/v)</li> |
| − | + | <li>50 mM Sodium acetate buffer, pH 5.0</li> | |
| − | <p>- | + | <li>100 mM Pyrophosphate</li> |
| − | <p> | + | <li>1 M Citric acid</li> |
| − | <p>- | + | <li>Protein sample</li> |
| − | + | </ul> | |
| + | <p><strong>Procedure:</strong></p> | ||
| + | <ul> | ||
| + | <li>Set a heating block to 37°C or 40°C</li> | ||
| + | <li>Place 2 mL reaction tubes for each sample (technical triplicates) plus one blank in the heating block</li> | ||
| + | <li>Pipet 250 µL of 50mM Sodium acetate buffer in all reaction tubes</li> | ||
| + | <li>Add 40 µL of protein sample</li> | ||
| + | <li>Add 10 µL of 100 mM Pyrophosphate to the tubes</li> | ||
| + | <li>Mix the tubes by inverting</li> | ||
| + | <li>Incubate on the heating block for 30 minutes</li> | ||
| + | <li>Add 1.5 mL of AAM solution and homogenize by inverting the tubes</li> | ||
| + | <li>Add 100 µL 1 M Citric acid</li> | ||
| + | <li>If phosphatase is present, the colour will change from white/transparent to yellow</li> | ||
| + | <li>Measure the colour strength in a photometer at 355 nm</li> | ||
| + | </ul> | ||
| + | <p>CAUTION: Make sure that the buffer you used for protein extraction does not contain any phosphate components. Otherwise the test will be false positive!</p> | ||
| + | </div> | ||
| + | |||
| + | <a href="" onClick=" $('#menu13').slideToggle(300, function callback() { }); return false;"> | ||
| + | <h1>Cellulase activity screening</h1></a> | ||
| + | <div id="menu13"> | ||
| + | |||
| + | <p align="center"><span style="font-size: x-large; color: #888888;"><strong></strong></span></p> | ||
| + | <p align="center"> </p> | ||
| + | <ul> | ||
| + | <li>Streak Competent Top10 <em>E. coli</em> cells or Competent BL21 cells transformed with appropriate vector containing cellulase construct onto agar plates containing cellulose substrate.</li> | ||
| + | <li>Incubate plates at 37<sup>o</sup>C for minimum 3-4 days (timing can be extended depending on bacterial strain).</li> | ||
| + | <li>After 3-4 days look for plates having clear zone of activity around cellulose substrate.</li> | ||
| + | </div> | ||
| + | |||
| + | <a href="" onClick=" $('#menu41').slideToggle(300, function callback() { }); return false;"><h1 style="color:white;"> Enzymatic Activity Test for Cellulase </h1></a> | ||
| + | <div id="menu41"> | ||
| + | |||
| + | |||
| + | <p> | ||
| + | Solutions: | ||
| + | </p> | ||
| + | <p> | ||
| + | Buffer: | ||
| + | </p> | ||
| + | <p> | ||
| + | Tris (MW: 121.14 g/mol) 50 mM | ||
| + | </p> | ||
| + | <p> | ||
| + | dH<sub>2</sub>O ad 1 l | ||
| + | </p> | ||
| + | <p> | ||
| + | pH 7 | ||
| + | </p> | ||
| + | <p> | ||
| + | Substrate solution: | ||
| + | </p> | ||
| + | <p> | ||
| + | Carboxymethil cellulose 1 % (w / v) | ||
| + | </p> | ||
| + | <p> | ||
| + | ddH<sub>2</sub>O ad 1 l | ||
| + | </p> | ||
| + | <p> | ||
| + | DNSA reagent solution: | ||
| + | </p> | ||
| + | <p> | ||
| + | Dinitrosalycylic acid 10 g | ||
| + | </p> | ||
| + | <p> | ||
| + | Phenol 2 ml | ||
| + | </p> | ||
| + | <p> | ||
| + | K Na Tartrate 200 g | ||
| + | </p> | ||
| + | <p> | ||
| + | NaOH 10 g | ||
| + | </p> | ||
| + | <p> | ||
| + | Na<sub>2</sub>SO<sub>3</sub> 0.5 g | ||
| + | </p> | ||
| + | <p> | ||
| + | dH<sub>2</sub>O ad 1 l | ||
| + | </p> | ||
| + | <p> | ||
| + | Store at 4°C, protected from light | ||
| + | </p> | ||
| + | <p> | ||
| + | Procedure: | ||
| + | </p> | ||
| + | <p> | ||
| + | 250 µl substrate solution | ||
| + | </p> | ||
| + | <p> | ||
| + | 100 µl Tris-Buffer | ||
| + | </p> | ||
| + | <p> | ||
| + | x µl enzyme solution | ||
| + | </p> | ||
| + | <p> | ||
| + | ad 500 µl ddH<sub>2</sub>O | ||
| + | </p> | ||
| + | <p> | ||
| + | Incubate setup as described above at 37°C for 20 min. | ||
| + | </p> | ||
| + | <p> | ||
| + | Add 750 µl DNSA. Heat the setup at 96°C for 15 min. | ||
| + | </p> | ||
| + | <p> | ||
| + | Put mixture on ice, centrifuge shortly at 4°C, 13000 x g. | ||
| + | </p> | ||
| + | <p> | ||
| + | Measure OD<sub>575</sub> against same setup without enzyme as a blank. | ||
| + | </p> | ||
| + | </div> | ||
| + | |||
| + | <h2> Restriction Controls </h2> | ||
| + | <a href="" onClick=" $('#menu17').slideToggle(300, function callback() { }); return false;"><h1> Aan I (Psi I ) - thermo fisher scientific - restriction control protocol</h1></a> | ||
| + | <div id="menu17"> | ||
| + | <p>- Add the following components</p> | ||
| + | <table border="1" cellspacing="0" cellpadding="0"> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td valign="top" width="151"> | ||
| + | <p>Purified Plasmid</p> | ||
| + | </td> | ||
| + | <td valign="top" width="126"> | ||
| + | <p>1 µg</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="151"> | ||
| + | <p>10X Buffer Tango</p> | ||
| + | </td> | ||
| + | <td valign="top" width="126"> | ||
| + | <p>1 µl</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="151"> | ||
| + | <p>Ana I (Psi I) (10 U/L)</p> | ||
| + | </td> | ||
| + | <td valign="top" width="126"> | ||
| + | <p>0.5 µl</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="151"> | ||
| + | <p>Nuclease free H<sub>2</sub>O</p> | ||
| + | </td> | ||
| + | <td valign="top" width="126"> | ||
| + | <p>Add to 10 µl</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | <p>- Incubate at 37 <sup>o</sup>C for 2 hours</p> | ||
| + | <p>- For inactivation incubation at 65°C for 20 min. </p> <p></p> | ||
<p> </p> | <p> </p> | ||
<p></p> | <p></p> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<p> </p> | <p> </p> | ||
</div> | </div> | ||
| − | + | ||
| − | <div id=" | + | |
| − | + | <a href="" onClick=" $('#menu18').slideToggle(300, function callback() { }); return false;"> | |
| − | + | <h1> Double digestion restriction control</h1></a> | |
| − | + | <div id="menu18"> | |
| − | + | ||
| − | + | <p align="center"><span style="font-size: x-large; color: #888888;"><strong></strong></span></p> | |
| − | + | <p align="center"> </p> | |
| − | <table border="1" cellspacing="0" cellpadding="0"> | + | <ul> |
| − | + | <p align="center"><strong>Double digestion restriction control</strong></p> | |
| + | <p><strong> </strong></p> | ||
| + | <p><strong>Sac I and Pvu II - thermo fisher scientific - restriction control protocol</strong> </p> | ||
| + | <p> </p> | ||
| + | <ul> | ||
| + | <li>Add the following components </li> | ||
| + | </ul> | ||
| + | <table style="width: 254px;" border="1" cellspacing="0" cellpadding="0" align="left"> | ||
| + | <tbody> | ||
<tr> | <tr> | ||
<td valign="top"> | <td valign="top"> | ||
| − | <p> | + | <p>Purified Plasmid </p> |
</td> | </td> | ||
<td valign="top"> | <td valign="top"> | ||
| − | <p> | + | <p>800 ng</p> |
</td> | </td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td valign="top"> | <td valign="top"> | ||
| − | <p> | + | <p>Sac I (2x)</p> |
</td> | </td> | ||
<td valign="top"> | <td valign="top"> | ||
| − | <p> | + | <p>2 µl </p> |
</td> | </td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td valign="top"> | <td valign="top"> | ||
| − | <p> | + | <p>Pvu II (1x)</p> |
</td> | </td> | ||
<td valign="top"> | <td valign="top"> | ||
| − | <p> | + | <p>1 µl </p> |
</td> | </td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td valign="top"> | <td valign="top"> | ||
| − | <p> | + | <p>10x Tango buffer</p> |
</td> | </td> | ||
<td valign="top"> | <td valign="top"> | ||
| − | <p> | + | <p>2 µl </p> |
</td> | </td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td valign="top"> | <td valign="top"> | ||
| − | <p> | + | <p>Nuclease free H<sub>2</sub>O </p> |
</td> | </td> | ||
<td valign="top"> | <td valign="top"> | ||
| − | <p> | + | <p>Add to 20 µl </p> |
</td> | </td> | ||
</tr> | </tr> | ||
| − | + | </tbody> | |
| − | + | </table> | |
| − | + | <p> </p> | |
| − | + | <p> </p> | |
| − | + | <p> </p> | |
| − | + | <p> </p> | |
| − | </ul> | + | <p align="center"> </p> |
| − | + | <p> </p> | |
| − | + | <p> </p> | |
| − | + | <p> </p> | |
| − | <p> | + | <p> </p> |
| − | + | <p> </p> | |
| − | + | <p> </p> | |
| − | + | <p></p> | |
| − | + | <ul> | |
| − | + | <li>For 10 µl reaction add 1 µl of 10x Tango buffer, for 20 µl reaction add 2 µl of 10x Tango buffer and vice versa.</li> | |
| − | + | <li>Incubate at 37 <sup>o</sup>C for 2 hours.</li> | |
| − | + | <li>For inactivation incubation at 80°C for 5 min. </li> | |
| − | <p>< | + | <li>Check the products on 0.8% agarose gel.</li> |
| − | + | </ul> | |
| − | + | </div> | |
| + | |||
| + | <a href="" onClick=" $('#menu19').slideToggle(300, function callback() { }); return false;"><h1> Restriction control using fast and slow digestion enzymes</h1></a> | ||
| + | <div id="menu19"> | ||
| + | <p align="center"><strong>Restriction control using fast digestion enzymes</strong></p> | ||
| + | <p><strong> </strong></p> | ||
| + | <p><strong>Sac I and HindIII - thermo fisher scientific - restriction control protocol</strong> </p> | ||
| + | <p> </p> | ||
| + | <ul> | ||
| + | <li>Add the following components </li> | ||
| + | </ul> | ||
| + | <table style="width: 223px;" border="1" cellspacing="0" cellpadding="0" align="left"> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td valign="top" width="127"> | ||
| + | <p>Purified Plasmid </p> | ||
| + | </td> | ||
| + | <td valign="top" width="96"> | ||
| + | <p>500-1,000 ng</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="127"> | ||
| + | <p>Sac I </p> | ||
| + | </td> | ||
| + | <td valign="top" width="96"> | ||
| + | <p>1 µl </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="127"> | ||
| + | <p>Hind III </p> | ||
| + | </td> | ||
| + | <td valign="top" width="96"> | ||
| + | <p>1 µl </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="127"> | ||
| + | <p>Nuclease free H<sub>2</sub>O </p> | ||
| + | </td> | ||
| + | <td valign="top" width="96"> | ||
| + | <p>Add to 10 µl </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <ul> | ||
| + | <li>Incubate at 37 <sup>o</sup>C for 30 min.</li> | ||
| + | <li>For inactivation incubation at 65°C for 5 min. </li> | ||
| + | </ul> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p><strong>PST I - thermo fisher scientific - restriction control protocol</strong> </p> | ||
| + | <p> </p> | ||
| + | <ul> | ||
| + | <li>Add the following components </li> | ||
| + | </ul> | ||
| + | <table style="width: 254px;" border="1" cellspacing="0" cellpadding="0" align="left"> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td valign="top"> | ||
| + | <p>Purified Plasmid </p> | ||
| + | </td> | ||
| + | <td valign="top"> | ||
| + | <p>500-1,000 ng</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top"> | ||
| + | <p>FD buffer</p> | ||
| + | </td> | ||
| + | <td valign="top"> | ||
| + | <p>1 µl </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top"> | ||
| + | <p>PST I </p> | ||
| + | </td> | ||
| + | <td valign="top"> | ||
| + | <p>1 µl </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top"> | ||
| + | <p>Nuclease free H<sub>2</sub>O </p> | ||
| + | </td> | ||
| + | <td valign="top"> | ||
| + | <p>Add to 10 µl </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <ul> | ||
| + | <li>Incubate at 37 <sup>o</sup>C for 30 min.</li> | ||
| + | <li>Products were checked on 0.8% agarose gel.</li> | ||
| + | </ul> | ||
| + | <p> </p> | ||
| + | <p align="center"><strong>Restriction control using slow digestion enzymes</strong></p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p><strong>Bsm I - fermentas - restriction control protocol</strong> </p> | ||
| + | <p> </p> | ||
| + | <ul> | ||
| + | <li>Add the following components </li> | ||
| + | </ul> | ||
| + | <table style="width: 254px;" border="1" cellspacing="0" cellpadding="0" align="left"> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td valign="top"> | ||
| + | <p>Purified Plasmid </p> | ||
| + | </td> | ||
| + | <td valign="top"> | ||
| + | <p>500 ng </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top"> | ||
| + | <p>Red buffer</p> | ||
| + | </td> | ||
| + | <td valign="top"> | ||
| + | <p>1 µl </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top"> | ||
| + | <p>Bsm I </p> | ||
| + | </td> | ||
| + | <td valign="top"> | ||
| + | <p>1 µl </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top"> | ||
| + | <p>Nuclease free H<sub>2</sub>O </p> | ||
| + | </td> | ||
| + | <td valign="top"> | ||
| + | <p>Add to 8 µl </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <ul> | ||
| + | <li>Incubate at 37 <sup>o</sup>C for 3 hours.</li> | ||
| + | <li>Products were checked on 0.8% agarose gel.</li> | ||
| + | </ul> | ||
| + | <p> </p> | ||
| + | <p></p> | ||
<p> </p> | <p> </p> | ||
| + | <p></p> | ||
<p> </p> | <p> </p> | ||
</div> | </div> | ||
| + | |||
| − | <a href="" onClick=" $('# | + | <a href="" onClick=" $('#menu20').slideToggle(300, function callback() { }); return false;"> |
| − | <h1> | + | <h1> Scafoldin Restriction control </h1></a> |
| − | <div id=" | + | <div id="menu20"> |
<p align="center"><span style="font-size: x-large; color: #888888;"><strong></strong></span></p> | <p align="center"><span style="font-size: x-large; color: #888888;"><strong></strong></span></p> | ||
<p align="center"> </p> | <p align="center"> </p> | ||
<ul> | <ul> | ||
| − | < | + | <p><strong>Scaffoldin Restriction Control using the restriction enzyme BsmI</strong></p> |
| − | < | + | <p>Set up the reaction as follows,</p> |
| − | + | <table border="1" cellspacing="0" cellpadding="0"> | |
| − | </ | + | <tbody> |
| + | <tr> | ||
| + | <td valign="top" width="301"> | ||
| + | <p>Components</p> | ||
| + | </td> | ||
| + | <td valign="top" width="301"> | ||
| + | <p>Quantity</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="301"> | ||
| + | <p>Nuclease free Water</p> | ||
| + | </td> | ||
| + | <td valign="top" width="301"> | ||
| + | <p>15 µl</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="301"> | ||
| + | <p>Red Buffer 10X</p> | ||
| + | </td> | ||
| + | <td valign="top" width="301"> | ||
| + | <p>2 µl</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="301"> | ||
| + | <p>DNA</p> | ||
| + | </td> | ||
| + | <td valign="top" width="301"> | ||
| + | <p>1,000 ng (2 µl)</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="301"> | ||
| + | <p>BsmI</p> | ||
| + | </td> | ||
| + | <td valign="top" width="301"> | ||
| + | <p>1 µl</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="301"> | ||
| + | <p> </p> | ||
| + | </td> | ||
| + | <td valign="top" width="301"> | ||
| + | <p>20 µl</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | |||
| − | |||
| − | |||
| − | |||
<p> </p> | <p> </p> | ||
| − | <p | + | <p>Incubate the mixture for 2 hours at 37ᵒC.</p> |
| − | + | <p>Perform the heat inactivation step for the reaction mixture for 10 min at 65ᵒC.</p> | |
| − | + | ||
| − | + | ||
| − | <p> | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</div> | </div> | ||
| + | |||
| + | <a href="" onClick=" $('#menu21').slideToggle(300, function callback() { }); return false;"><h1> Esterase Restriction Control</h1></a> | ||
| + | <div id="menu21"> | ||
| + | <p align="center"><strong>Esterase Restriction Control</strong></p> | ||
<p> </p> | <p> </p> | ||
| − | < | + | <table style="width: 251px;" border="0" cellspacing="0" cellpadding="0"> |
| − | <p | + | <tbody> |
| + | <tr> | ||
| + | <td valign="top"> | ||
| + | <p>Nuclease Free Water</p> | ||
| + | </td> | ||
| + | <td valign="top"> | ||
| + | <p>15 µl</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top"> | ||
| + | <p>R Buffer 10X</p> | ||
| + | </td> | ||
| + | <td valign="top"> | ||
| + | <p>2 µl</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top"> | ||
| + | <p>DNA (up to 1µg)</p> | ||
| + | </td> | ||
| + | <td valign="top"> | ||
| + | <p>2 µl</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top"> | ||
| + | <p>XhoI (10U/µl)</p> | ||
| + | </td> | ||
| + | <td valign="top"> | ||
| + | <p>1 µl</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top"> | ||
| + | <p>PstI (10U/µl)</p> | ||
| + | </td> | ||
| + | <td valign="top"> | ||
| + | <p>1 µl</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
<p> </p> | <p> </p> | ||
| − | <p> | + | <p>Incubate 20 min at 37˚C and inactivate 5 min at 80˚C.</p> |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
<p> </p> | <p> </p> | ||
| − | <p><span style="text- | + | <p></p> |
| − | <p> | + | <p> </p> |
| − | <p>3, | + | <p></p> |
| − | <p> | + | <p> </p> |
| − | <p> | + | </div> |
| − | <p> | + | |
| − | <p> | + | |
| − | <p> | + | <a href="" onClick=" $('#menu22').slideToggle(300, function callback() { }); return false;"> |
| − | <p> | + | <h1> Phosphatase Restriction Control</h1></a> |
| + | <div id="menu22"> | ||
| + | |||
| + | <p align="center"><span style="font-size: x-large; color: #888888;"><strong></strong></span></p> | ||
| + | <p align="center"> </p> | ||
| + | <ul> | ||
| + | <p class="MsoNormal" style="margin-bottom: 0.0001pt; text-align: center;" align="center"><strong><span style="font-size: 10pt; font-family: Arial, sans-serif;" lang="ES-CR">Phosphatase Restriction Control</span></strong></p> | ||
| + | <p class="MsoNormal" style="margin-bottom: 0.0001pt;"><span lang="ES-CR"> </span></p> | ||
| + | <table class="MsoNormalTable" style="width: 288px; border-collapse: collapse; margin-left: 4.8pt; margin-right: 4.8pt;" border="0" cellspacing="0" cellpadding="0" align="left"> | ||
| + | <tbody> | ||
| + | <tr style="mso-yfti-irow: 0; mso-yfti-firstrow: yes; height: 12.15pt;"> | ||
| + | <td style="border: solid black 1.0pt; mso-border-alt: solid black .75pt; padding: 4.75pt 4.75pt 4.75pt 4.75pt; height: 12.15pt;" valign="top"> | ||
| + | <p class="MsoNormal" style="margin-bottom: .0001pt; mso-line-height-alt: 0pt; mso-element: frame; mso-element-frame-hspace: 7.05pt; mso-element-wrap: around; mso-element-anchor-vertical: paragraph; mso-element-anchor-horizontal: page; mso-element-left: 128.4pt; mso-element-top: 3.8pt; mso-height-rule: exactly;"><span style="font-size: 10pt; font-family: Arial, sans-serif;" lang="ES-CR">Nuclease Free Water</span></p> | ||
| + | </td> | ||
| + | <td style="border: solid black 1.0pt; border-left: none; mso-border-left-alt: solid black .75pt; mso-border-alt: solid black .75pt; padding: 4.75pt 4.75pt 4.75pt 4.75pt; height: 12.15pt;" valign="top"> | ||
| + | <p class="MsoNormal" style="margin-top: 0in; margin-right: 0in; margin-bottom: .0001pt; margin-left: 35.4pt; text-indent: -35.4pt; mso-line-height-alt: 0pt; mso-element: frame; mso-element-frame-hspace: 7.05pt; mso-element-wrap: around; mso-element-anchor-vertical: paragraph; mso-element-anchor-horizontal: page; mso-element-left: 128.4pt; mso-element-top: 3.8pt; mso-height-rule: exactly;"><span style="font-size: 10pt; font-family: Arial, sans-serif;" lang="ES-CR">14µl</span></p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr style="mso-yfti-irow: 1; height: 12.15pt;"> | ||
| + | <td style="border: solid black 1.0pt; border-top: none; mso-border-top-alt: solid black .75pt; mso-border-alt: solid black .75pt; padding: 4.75pt 4.75pt 4.75pt 4.75pt; height: 12.15pt;" valign="top"> | ||
| + | <p class="MsoNormal" style="margin-bottom: .0001pt; mso-line-height-alt: 0pt; mso-element: frame; mso-element-frame-hspace: 7.05pt; mso-element-wrap: around; mso-element-anchor-vertical: paragraph; mso-element-anchor-horizontal: page; mso-element-left: 128.4pt; mso-element-top: 3.8pt; mso-height-rule: exactly;"><span style="font-size: 10pt; font-family: Arial, sans-serif;" lang="ES-CR">Fast digestion Buffer 10X</span></p> | ||
| + | </td> | ||
| + | <td style="border-top: none; border-left: none; border-bottom: solid black 1.0pt; border-right: solid black 1.0pt; mso-border-top-alt: solid black .75pt; mso-border-left-alt: solid black .75pt; mso-border-alt: solid black .75pt; padding: 4.75pt 4.75pt 4.75pt 4.75pt; height: 12.15pt;" valign="top"> | ||
| + | <p class="MsoNormal" style="margin-bottom: .0001pt; mso-line-height-alt: 0pt; mso-element: frame; mso-element-frame-hspace: 7.05pt; mso-element-wrap: around; mso-element-anchor-vertical: paragraph; mso-element-anchor-horizontal: page; mso-element-left: 128.4pt; mso-element-top: 3.8pt; mso-height-rule: exactly;"><span style="font-size: 10pt; font-family: Arial, sans-serif;" lang="ES-CR">2µl</span></p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr style="mso-yfti-irow: 2; height: 12.15pt;"> | ||
| + | <td style="border: solid black 1.0pt; border-top: none; mso-border-top-alt: solid black .75pt; mso-border-alt: solid black .75pt; padding: 4.75pt 4.75pt 4.75pt 4.75pt; height: 12.15pt;" valign="top"> | ||
| + | <p class="MsoNormal" style="margin-bottom: .0001pt; mso-line-height-alt: 0pt; mso-element: frame; mso-element-frame-hspace: 7.05pt; mso-element-wrap: around; mso-element-anchor-vertical: paragraph; mso-element-anchor-horizontal: page; mso-element-left: 128.4pt; mso-element-top: 3.8pt; mso-height-rule: exactly;"><span style="font-size: 10pt; font-family: Arial, sans-serif;" lang="ES-CR">DNA ( up to 1µg)</span></p> | ||
| + | </td> | ||
| + | <td style="border-top: none; border-left: none; border-bottom: solid black 1.0pt; border-right: solid black 1.0pt; mso-border-top-alt: solid black .75pt; mso-border-left-alt: solid black .75pt; mso-border-alt: solid black .75pt; padding: 4.75pt 4.75pt 4.75pt 4.75pt; height: 12.15pt;" valign="top"> | ||
| + | <p class="MsoNormal" style="margin-bottom: .0001pt; mso-line-height-alt: 0pt; mso-element: frame; mso-element-frame-hspace: 7.05pt; mso-element-wrap: around; mso-element-anchor-vertical: paragraph; mso-element-anchor-horizontal: page; mso-element-left: 128.4pt; mso-element-top: 3.8pt; mso-height-rule: exactly;"><span style="font-size: 10pt; font-family: Arial, sans-serif;" lang="ES-CR">2µl</span></p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr style="mso-yfti-irow: 3; height: 12.8pt;"> | ||
| + | <td style="border: solid black 1.0pt; border-top: none; mso-border-top-alt: solid black .75pt; mso-border-alt: solid black .75pt; padding: 4.75pt 4.75pt 4.75pt 4.75pt; height: 12.8pt;" valign="top"> | ||
| + | <p class="MsoNormal" style="margin-bottom: .0001pt; mso-line-height-alt: 0pt; mso-element: frame; mso-element-frame-hspace: 7.05pt; mso-element-wrap: around; mso-element-anchor-vertical: paragraph; mso-element-anchor-horizontal: page; mso-element-left: 128.4pt; mso-element-top: 3.8pt; mso-height-rule: exactly;"><span style="font-size: 10pt; font-family: Arial, sans-serif;" lang="ES-CR">SacI (10U/µl)</span></p> | ||
| + | </td> | ||
| + | <td style="border-top: none; border-left: none; border-bottom: solid black 1.0pt; border-right: solid black 1.0pt; mso-border-top-alt: solid black .75pt; mso-border-left-alt: solid black .75pt; mso-border-alt: solid black .75pt; padding: 4.75pt 4.75pt 4.75pt 4.75pt; height: 12.8pt;" valign="top"> | ||
| + | <p class="MsoNormal" style="margin-bottom: .0001pt; mso-line-height-alt: 0pt; mso-element: frame; mso-element-frame-hspace: 7.05pt; mso-element-wrap: around; mso-element-anchor-vertical: paragraph; mso-element-anchor-horizontal: page; mso-element-left: 128.4pt; mso-element-top: 3.8pt; mso-height-rule: exactly;"><span style="font-size: 10pt; font-family: Arial, sans-serif;" lang="ES-CR">1µl</span></p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr style="mso-yfti-irow: 4; mso-yfti-lastrow: yes; height: 12.15pt;"> | ||
| + | <td style="border: solid black 1.0pt; border-top: none; mso-border-top-alt: solid black .75pt; mso-border-alt: solid black .75pt; padding: 4.75pt 4.75pt 4.75pt 4.75pt; height: 12.15pt;" valign="top"> | ||
| + | <p class="MsoNormal" style="margin-bottom: .0001pt; mso-line-height-alt: 0pt; mso-element: frame; mso-element-frame-hspace: 7.05pt; mso-element-wrap: around; mso-element-anchor-vertical: paragraph; mso-element-anchor-horizontal: page; mso-element-left: 128.4pt; mso-element-top: 3.8pt; mso-height-rule: exactly;"><span style="font-size: 10pt; font-family: Arial, sans-serif;" lang="ES-CR">KpnI(10U/µl)</span></p> | ||
| + | </td> | ||
| + | <td style="border-top: none; border-left: none; border-bottom: solid black 1.0pt; border-right: solid black 1.0pt; mso-border-top-alt: solid black .75pt; mso-border-left-alt: solid black .75pt; mso-border-alt: solid black .75pt; padding: 4.75pt 4.75pt 4.75pt 4.75pt; height: 12.15pt;" valign="top"> | ||
| + | <p class="MsoNormal" style="margin-bottom: .0001pt; mso-line-height-alt: 0pt; mso-element: frame; mso-element-frame-hspace: 7.05pt; mso-element-wrap: around; mso-element-anchor-vertical: paragraph; mso-element-anchor-horizontal: page; mso-element-left: 128.4pt; mso-element-top: 3.8pt; mso-height-rule: exactly;"><span style="font-size: 10pt; font-family: Arial, sans-serif;" lang="ES-CR">1µl</span></p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | <p class="MsoNormal" style="margin-bottom: 0.0001pt;"><span lang="ES-CR"> </span></p> | ||
| + | <p class="MsoNormal" style="margin-bottom: 0.0001pt;"><span lang="ES-CR"> </span></p> | ||
| + | <p class="MsoNormal" style="margin-bottom: 0.0001pt;"><span lang="ES-CR"> </span></p> | ||
| + | <p class="MsoNormal" style="margin-bottom: 0.0001pt;"><span lang="ES-CR"> </span></p> | ||
| + | <p class="MsoNormal" style="margin-bottom: 0.0001pt;"><span lang="ES-CR"> </span></p> | ||
| + | <p class="MsoNormal" style="margin-bottom: 0.0001pt;"><span lang="ES-CR"> </span></p> | ||
| + | <p class="MsoNormal" style="margin-bottom: 0.0001pt;"><span lang="ES-CR"> </span></p> | ||
| + | <p class="MsoNormal" style="margin-bottom: 0.0001pt;"><span lang="ES-CR"> </span></p> | ||
| + | <p class="MsoNormal" style="margin-bottom: 0.0001pt;"><span lang="ES-CR"> </span></p> | ||
| + | <p class="MsoNormal" style="margin-bottom: 0.0001pt;"><span lang="ES-CR"> </span></p> | ||
| + | <p class="MsoNormal" style="margin-bottom: 0.0001pt;"><span style="font-size: 10pt; font-family: Arial, sans-serif;" lang="EN-GB">Incubate 2 hours at 37˚C and inactivate 20min at 80˚C.</span></p> | ||
| + | <p align="center"> </p> | ||
| + | <p class="MsoNormal"><span style="mso-bidi-font-size: 10.0pt; line-height: 115%; mso-ansi-language: EN-GB;" lang="EN-GB"> </span></p> | ||
| + | </div> | ||
| + | |||
| + | <h2> PCR Preparation Methods </h2> | ||
| + | <a href="" onClick=" $('#menu23').slideToggle(300, function callback() { }); return false;"><h1> Colony PCR</h1></a> | ||
| + | <div id="menu23"> | ||
| + | <p align="center"><strong>Colony PCR</strong></p> | ||
<p> </p> | <p> </p> | ||
| − | <p> | + | <p>- Pick a colony with a tip and re suspend it in 5ul LB</p> |
| + | <p>- Prepare the PCR mix as follows:</p> | ||
<p> </p> | <p> </p> | ||
| − | <p> | + | <div align="center"> |
| − | <p> | + | <table style="width: 243px;" border="0" cellspacing="0" cellpadding="0"> |
| − | <p> | + | <tbody> |
| − | <p> | + | <tr> |
| − | <p> | + | <td valign="top" width="186"> |
| − | <p> | + | <p>Water</p> |
| − | <p>< | + | </td> |
| + | <td valign="top" width="57"> | ||
| + | <p>12 ul</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="186"> | ||
| + | <p>Taq Buffer</p> | ||
| + | </td> | ||
| + | <td valign="top" width="57"> | ||
| + | <p>2 ul</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="186"> | ||
| + | <p>MgCl</p> | ||
| + | </td> | ||
| + | <td valign="top" width="57"> | ||
| + | <p>2 ul</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="186"> | ||
| + | <p>dNTPs</p> | ||
| + | </td> | ||
| + | <td valign="top" width="57"> | ||
| + | <p>0.5 µl</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="186"> | ||
| + | <p>Primer Forward</p> | ||
| + | </td> | ||
| + | <td valign="top" width="57"> | ||
| + | <p>1 ul</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="186"> | ||
| + | <p>Primer Reverse</p> | ||
| + | </td> | ||
| + | <td valign="top" width="57"> | ||
| + | <p>1 ul</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="186"> | ||
| + | <p>Taq Polymerase</p> | ||
| + | </td> | ||
| + | <td valign="top" width="57"> | ||
| + | <p>1.5 µl</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | </div> | ||
<p> </p> | <p> </p> | ||
| + | <p><span style="text-decoration: underline;"> </span></p> | ||
| + | <p>- <span style="text-decoration: underline;">Program: </span></p> | ||
| + | <p>Initial denaturalization: 98˚C, 5 min</p> | ||
| + | <p>Denaturalization: 96˚C, 45 s</p> | ||
| + | <p>Annealing: 70˚C, 30 s</p> | ||
| + | <p>Elongation: 72˚C, 25 s</p> | ||
| + | <p></p> | ||
| + | <p> </p> | ||
| + | <p></p> | ||
| + | <p> </p> | ||
| + | </div> | ||
| + | |||
| − | <a href="" onClick=" $('# | + | <a href="" onClick=" $('#menu24').slideToggle(300, function callback() { }); return false;"> |
| − | <h1> | + | <h1>Phusion PCR</h1></a> |
| − | <div id=" | + | <div id="menu24"> |
| − | <p> | + | |
| − | <p> | + | <p align="center"><span style="font-size: x-large; color: #888888;"><strong></strong></span></p> |
| + | <p align="center"> </p> | ||
| + | <ul> | ||
| + | <p>This protocol describes PCR procedure using Phusion High-Fidelity DNA Polymerase. <strong>(Thermo Fisher Scientific).</strong></p> | ||
| + | <p>• Note: the annealing conditions depend on the applied DNA polymerases (such as Taq DNA polymerases).</p> | ||
| + | <p>•Use 15–30 s/kb for extension. Do not exceed 1 min/kb.</p> | ||
| + | <p>•Phusion DNA Polymerases produce blunt end DNA products.</p> | ||
| + | <p>•All components and reaction mix are placed on ice.</p> | ||
| + | <p>- add the following components, mix and spin down.</p> | ||
| + | <table style="width: 428px;" border="1" cellspacing="0" cellpadding="0"> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td valign="top" nowrap="nowrap" width="130"> | ||
| + | <p><strong>Component</strong></p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="114"> | ||
| + | <p align="center"><strong>20 µL rxn</strong></p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="96"> | ||
| + | <p align="center"><strong>50 µL rxn</strong></p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="88"> | ||
| + | <p align="center"><strong>Final conc.</strong></p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" nowrap="nowrap" width="130"> | ||
| + | <p align="center">H<sub>2</sub>O</p> | ||
| + | </td> | ||
| + | <td valign="top" width="114"> | ||
| + | <p align="center">add to 20 µL</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="96"> | ||
| + | <p align="center">add to 50 µL</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="88"> </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="130"> | ||
| + | <p align="center">5X Phusion HF Buffer</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="114"> | ||
| + | <p align="center">4 µL</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="96"> | ||
| + | <p align="center">10 µL</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="88"> | ||
| + | <p align="center">1X</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" nowrap="nowrap" width="130"> | ||
| + | <p align="center">10 mM dNTPs</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="114"> | ||
| + | <p align="center">0.4 µL</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="96"> | ||
| + | <p align="center">1 µL</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="88"> | ||
| + | <p align="center">200 µM each</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" nowrap="nowrap" width="130"> | ||
| + | <p align="center">Forward primer</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="114"> | ||
| + | <p align="center">X µL</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="96"> | ||
| + | <p align="center">X µL</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="88"> | ||
| + | <p align="center">0.5 µM</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" nowrap="nowrap" width="130"> | ||
| + | <p align="center">Reverse primer</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="114"> | ||
| + | <p align="center">X µL</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="96"> | ||
| + | <p align="center">X µL</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="88"> | ||
| + | <p align="center">0.5 µM</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" nowrap="nowrap" width="130"> | ||
| + | <p align="center">Template DNA</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="114"> | ||
| + | <p align="center">15 -20 ng</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="96"> | ||
| + | <p align="center">30 - 50 ng</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="88"> </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" nowrap="nowrap" width="130"> | ||
| + | <p align="center">(DMSO, optional)*</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="114"> | ||
| + | <p align="center">(0.6 µL)</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="96"> | ||
| + | <p align="center">(1.5 µL)</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="88"> | ||
| + | <p align="center">3%</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="130"> | ||
| + | <p align="center">Phusion DNA Polymerase</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="114"> | ||
| + | <p align="center">0.2 µL</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="96"> | ||
| + | <p align="center">0.5 µL</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="88"> | ||
| + | <p align="center">0.02 U/µL</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
<p> </p> | <p> </p> | ||
| − | <p> | + | <p>* Addition of DMSO is recommended for GC-rich amplicons. DMSO is not recommended for amplicons with very low GC % or amplicons that are > 20 kb.</p> |
| − | <p> | + | <p>- adjust the thermocycle to the following isntructions and start the PCR.</p> |
| − | <p> | + | <table style="width: 428px;" border="1" cellspacing="0" cellpadding="0"> |
| − | <p> | + | <tbody> |
| − | <p> | + | <tr> |
| − | <p> | + | <td valign="top" nowrap="nowrap" width="130"> |
| − | <p> | + | <p><strong>Cycle step</strong></p> |
| − | <p> | + | </td> |
| − | <p> | + | <td valign="top" nowrap="nowrap" width="114"> |
| − | <p> | + | <p align="center"><strong>Tempreature <sup>o</sup>C</strong></p> |
| − | <p> | + | </td> |
| − | < | + | <td valign="top" nowrap="nowrap" width="96"> |
| − | < | + | <p align="center"><strong>Time </strong></p> |
| − | <p>< | + | </td> |
| − | <p>< | + | <td valign="top" nowrap="nowrap" width="88"> |
| − | <p>< | + | <p align="center"><strong>Cycles</strong></p> |
| − | <p> | + | </td> |
| − | <p> | + | </tr> |
| + | <tr> | ||
| + | <td valign="top" nowrap="nowrap" width="130"> | ||
| + | <p>Initial Denaturation</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="114"> | ||
| + | <p align="center">98</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="96"> | ||
| + | <p align="center">30 s</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="88"> | ||
| + | <p align="center">1</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" nowrap="nowrap" width="130"> | ||
| + | <p>Denaturation</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="114"> | ||
| + | <p align="center">98</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="96"> | ||
| + | <p align="center">5–10 s</p> | ||
| + | </td> | ||
| + | <td rowspan="3" valign="top" nowrap="nowrap" width="88"> | ||
| + | <p align="center">30 - 35</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" nowrap="nowrap" width="130"> | ||
| + | <p>Annealing</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="114"> | ||
| + | <p align="center">*</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="96"> | ||
| + | <p align="center">10–30 s</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" nowrap="nowrap" width="130"> | ||
| + | <p>Extension</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="114"> | ||
| + | <p align="center">72</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="96"> | ||
| + | <p align="center">15–30 s/kb</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" nowrap="nowrap" width="130"> | ||
| + | <p>Final extension</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="114"> | ||
| + | <p align="center">72</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="96"> | ||
| + | <p align="center">5-10 min</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="88"> | ||
| + | <p align="center">1</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
<p> </p> | <p> </p> | ||
| − | <table border="1" cellspacing="0" cellpadding="0"> | + | <p>* The optimal annealing temperature for Phusion DNA Polymerase may differ significantly from that of Taq-based polymerases. Always use the Tm calculator and instructions on our website (www.thermoscientific.com/pcrwebtools) to determine the Tm values of primers and optimal annealing temperature. The Phusion DNA Polymerase has the ability to stabilize primer-template hybridization. As a basic rule, for primers > 20 nt, anneal for 10–30 seconds at a Tm +3°C of the lower Tm primer. For primers ≤ 20 nt, use an annealing temperature equal to the Tm of the lower Tm primer.</p> |
| + | <p>- for Templates > 2.5 kb and primers with low stability apply the following program</p> | ||
| + | <table style="width: 428px;" border="1" cellspacing="0" cellpadding="0"> | ||
<tbody> | <tbody> | ||
<tr> | <tr> | ||
| − | <td> | + | <td valign="top" nowrap="nowrap" width="130"> |
| − | <p | + | <p><strong>Cycle step</strong></p> |
</td> | </td> | ||
| − | <td> | + | <td valign="top" nowrap="nowrap" width="114"> |
| − | <p align="center"> | + | <p align="center"><strong>Tempreature <sup>o</sup>C</strong></p> |
</td> | </td> | ||
| − | <td> | + | <td valign="top" nowrap="nowrap" width="96"> |
| − | <p align="center"> | + | <p align="center"><strong>Time </strong></p> |
</td> | </td> | ||
| − | <td> | + | <td valign="top" nowrap="nowrap" width="88"> |
| − | <p align="center"> | + | <p align="center"><strong>Cycles</strong></p> |
</td> | </td> | ||
| − | <td> | + | </tr> |
| − | <p align="center"> | + | <tr> |
| + | <td valign="top" nowrap="nowrap" width="130"> | ||
| + | <p>Initial Denaturation</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="114"> | ||
| + | <p align="center">98</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="96"> | ||
| + | <p align="center">30 s</p> | ||
</td> | </td> | ||
| − | <td> | + | <td valign="top" nowrap="nowrap" width="88"> |
| − | <p align="center"> | + | <p align="center">1</p> |
</td> | </td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td> | + | <td valign="top" nowrap="nowrap" width="130"> |
| − | <p | + | <p>Denaturation</p> |
</td> | </td> | ||
| − | <td> | + | <td valign="top" nowrap="nowrap" width="114"> |
| − | <p align="center"> | + | <p align="center">98</p> |
</td> | </td> | ||
| − | <td> | + | <td valign="top" nowrap="nowrap" width="96"> |
| − | <p align="center"> | + | <p align="center">10 s</p> |
</td> | </td> | ||
| − | <td> | + | <td rowspan="3" valign="top" nowrap="nowrap" width="88"> |
| − | <p align="center"> | + | <p align="center">30 - 35</p> |
</td> | </td> | ||
| − | <td> | + | </tr> |
| − | <p | + | <tr> |
| + | <td valign="top" nowrap="nowrap" width="130"> | ||
| + | <p>Annealing</p> | ||
</td> | </td> | ||
| − | <td> | + | <td valign="top" nowrap="nowrap" width="114"> |
| − | <p align="center"> | + | <p align="center">*</p> |
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="96"> | ||
| + | <p align="center">15 s</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" nowrap="nowrap" width="130"> | ||
| + | <p>Extension</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="114"> | ||
| + | <p align="center">72</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="96"> | ||
| + | <p align="center">1 min 30 s</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" nowrap="nowrap" width="130"> | ||
| + | <p>Final extension</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="114"> | ||
| + | <p align="center">72</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="96"> | ||
| + | <p align="center">10.5 min</p> | ||
| + | </td> | ||
| + | <td valign="top" nowrap="nowrap" width="88"> | ||
| + | <p align="center">1</p> | ||
</td> | </td> | ||
</tr> | </tr> | ||
</tbody> | </tbody> | ||
</table> | </table> | ||
| + | <p> </p> | ||
| + | <p>- check the PCR product on 0.8% agarose gel and store at - 20 celsius degrees.</p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
<p> </p> | <p> </p> | ||
</div> | </div> | ||
| − | |||
| − | < | + | <h2> Sequencing </h2> |
| − | + | <a href="" onClick=" $('#menu25').slideToggle(300, function callback() { }); return false;"><h1> Protocol for Sanger sequencing</h1></a> | |
| − | < | + | <div id="menu25"> |
<ul> | <ul> | ||
| − | <li> | + | <li>Sequencing reaction setup</li> |
| − | < | + | </ul> |
| − | <li> | + | <table style="width: 445px;" border="1" cellspacing="0" cellpadding="0" align="left"> |
| + | <tbody> | ||
| + | <tr> | ||
| + | <td valign="top" width="205"> | ||
| + | <p><strong>Template</strong></p> | ||
| + | </td> | ||
| + | <td valign="top" width="240"> | ||
| + | <p><strong>DNA amount</strong></p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="205"> | ||
| + | <p>PCR products </p> | ||
| + | </td> | ||
| + | <td valign="top" width="240"> | ||
| + | <p>10 ng/100 bp (if larger 1.5 kb 150 ng)</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="205"> | ||
| + | <p>Plasmids</p> | ||
| + | </td> | ||
| + | <td valign="top" width="240"> | ||
| + | <p>300 ng</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="205"> | ||
| + | <p>Forward primer (5pmol/ µl )</p> | ||
| + | </td> | ||
| + | <td valign="top" width="240"> | ||
| + | <p>1 µl</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="205"> | ||
| + | <p>Reverse primer (5pmol/ µl )</p> | ||
| + | </td> | ||
| + | <td valign="top" width="240"> | ||
| + | <p>1 µl</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="205"> | ||
| + | <p>Nuclease free H<sub>2</sub>O</p> | ||
| + | </td> | ||
| + | <td valign="top" width="240"> | ||
| + | <p>ad. 5 µl</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="205"> | ||
| + | <p><strong>Total volume</strong></p> | ||
| + | </td> | ||
| + | <td valign="top" width="240"> | ||
| + | <p><strong>5 </strong><strong>µl</strong></p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <ul> | ||
| + | <li>Forward and reverse primers should be added in 2 individual sample tubes!</li> | ||
</ul> | </ul> | ||
| + | </div> | ||
| + | |||
| + | <a href="" onClick=" $('#menu26').slideToggle(300, function callback() { }); return false;"> | ||
| + | <h1> Overnight Sanger Sequencing</h1></a> | ||
| + | <div id="menu26"> | ||
| − | < | + | <p align="center"><span style="font-size: x-large; color: #888888;"><strong></strong></span></p> |
| + | <p align="center"> </p> | ||
<ul> | <ul> | ||
| − | + | <p align="center"><strong>Sanger sequencing reaction setup for overnight</strong></p> | |
| − | + | <table style="width: 289px;" border="1" cellspacing="0" cellpadding="0" align="left"> | |
| − | + | <tbody> | |
| + | <tr> | ||
| + | <td valign="top" width="205"> | ||
| + | <p><strong>Components</strong></p> | ||
| + | </td> | ||
| + | <td valign="top" width="84"> | ||
| + | <p><strong>Volume</strong></p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="205"> | ||
| + | <p>Purified Plasmid </p> | ||
| + | </td> | ||
| + | <td valign="top" width="84"> | ||
| + | <p>800 ng</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="205"> | ||
| + | <p>Forward primer (1:10)</p> | ||
| + | </td> | ||
| + | <td valign="top" width="84"> | ||
| + | <p>3 µl</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="205"> | ||
| + | <p>Reverse primer (1:10)</p> | ||
| + | </td> | ||
| + | <td valign="top" width="84"> | ||
| + | <p>3 µl</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="205"> | ||
| + | <p>Nuclease free H<sub>2</sub>O</p> | ||
| + | </td> | ||
| + | <td valign="top" width="84"> | ||
| + | <p>ad. 15 µl</p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="205"> | ||
| + | <p><strong>Total volume</strong></p> | ||
| + | </td> | ||
| + | <td valign="top" width="84"> | ||
| + | <p><strong>15 </strong><strong>µl</strong></p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | |||
| + | |||
| + | <ul> | ||
| + | <li>Forward and reverse primers should be added in 2 individual sample tubes!</li> | ||
</ul> | </ul> | ||
</div> | </div> | ||
| + | |||
| + | <h2> Fluorescence Microscopy </h2> | ||
| + | <a href="" onClick=" $('#menu27').slideToggle(300, function callback() { }); return false;"><h1>RFP microscopy</h1></a> | ||
| + | <div id="menu27"> | ||
| + | <ul> | ||
| + | <p><strong>RFP microscopy</strong></p> | ||
| + | <p>To check if the transformed <em>E.coli</em> TOP10 show red fluorescence, the culture was examined by fluorescence microscopy. The RFP DsRed filter was used (excitation at 536 nm, emission at 582 nm). To prevent the cells from floating around they are fixed in 0.8%.</p> | ||
| + | <p> </p> | ||
| + | <p><strong>Preparation of slides</strong></p> | ||
| + | <ul> | ||
| + | <li>Prepare 0.8% agarose with water and boil it up.</li> | ||
| + | <li>Pipet 500 μL onto a slide and press another one on top. Let the agarose cure between both slides.</li> | ||
| + | <li>Carefully remove the upper slide and put the sample name in one corner the one with the agarose.</li> | ||
| + | <li>Dilute the desired cell cultures 1:10 up to 1:50 (depends on thickness) and remember to prepare also a positive (if available) and negative control.</li> | ||
| + | <li>Pipet 10-20 μL of the diluted culture on the agarose and put the cover slip on top.</li> | ||
| + | <li>Perform fluorescence microscopy with the right filter.</li> | ||
| + | <li>If successful, take pictures of the red fluorescing <em>E. coli</em>.</li> | ||
| + | <li>Note: Agarose has a slight self-fluorescence.</li> | ||
| + | </ul> | ||
| + | </div> | ||
| + | |||
| + | </html> | ||
| + | [[File:Counting_iGEM_Goettingen2015.jpeg|700px|thumb|center|]] | ||
| + | <html> | ||
| + | |||
| + | </div></div> <!--These are the closing tags for div id="mainContainer" and div id="contentContainer". The corresponding opening tags appear in the template that is {{included}} at the top of this page.--> | ||
</html> | </html> | ||
Latest revision as of 00:00, 19 September 2015
Media/Buffer
LB Medium
"Fat" LB Medium
Media and Culture Methods for Dockerin Organisms of Origin
Phosphatase Activity plates, Sperber media
Esterase Activity plates, with 1% Tributyrin
Cellulase activity plates
1x TAE Buffer
Cloning Methods
PCR product purification using QIAquick® PCR Purification Kit (QIAGEN)
PCR Gel extraction, peqGOLD Gel Extraction Kit
Blunt End Ligation in pJET1.2 vector –Clone JET PCR Cloning Kit– (Thermo Scientific)
Sticky End T4 Ligation (Thermo Scientific)
TOPO® Cloning protocol usingChampion™ pET Directional TOPO® Expression Kits (Thermo Fisher Scientific)
Plasmid transformation into chemically competent E. coli
Electroporation of BL21 cells with pJET_RFP
Plasmid Extraction - using QIAprep Spin Miniprep Kit (QIAGEN)
Plasmid Extraction - using peqGOLD Plasmid Miniprep Kit I (PEQLAB Technologies)
Competent Cells
Preparation of competent E.coli cells
Transformation Efficiency Kit, RFP construct (iGEM)
Protein Extraction and Purification
Induction, harvest and disruption of expression cultures
Protein Purification (Protino® Ni-IDA 2000 His-Tag protein purification, Macherey-Nagel)
Affinity chromatography of His-tagged proteins
Size Exclusion Chromatography
Concentration of protein solutions
Bradford Assay
SDS Polyacrylamid Gel Electrophoresis
Western Blot
Activity Screens & Tests
Esterase activity test
Phosphatase activity test
Cellulase activity screening
Enzymatic Activity Test for Cellulase
Restriction Controls
Aan I (Psi I ) - thermo fisher scientific - restriction control protocol
Double digestion restriction control
Restriction control using fast and slow digestion enzymes
Scafoldin Restriction control
Esterase Restriction Control
Phosphatase Restriction Control
PCR Preparation Methods
Colony PCR
Phusion PCR
Sequencing
Protocol for Sanger sequencing
Overnight Sanger Sequencing
Fluorescence Microscopy
RFP microscopy