Difference between revisions of "Team:Pretoria UP/Results"
| Line 22: | Line 22: | ||
#resultsDiv | #resultsDiv | ||
{ | { | ||
| − | width= | + | width=60%; |
} | } | ||
Revision as of 17:30, 17 September 2015
Project Results
Fluorescent Microscopy
The genetic switch systems that are being characterised make use of two different reporter genes, namely green fluorescent protein (GFP) and red fluorescent protein (RFP). These will give a visual indication as to the functioning of the parts being tested. The biobricks, Bba_I13521 and Bba_I13522 were transformed into E. coli and serve as positive controls for fluorescent microscopy. These two parts consist of the reporter genes, GFP and RFP respectively and are under the control of a constitutive promoter.
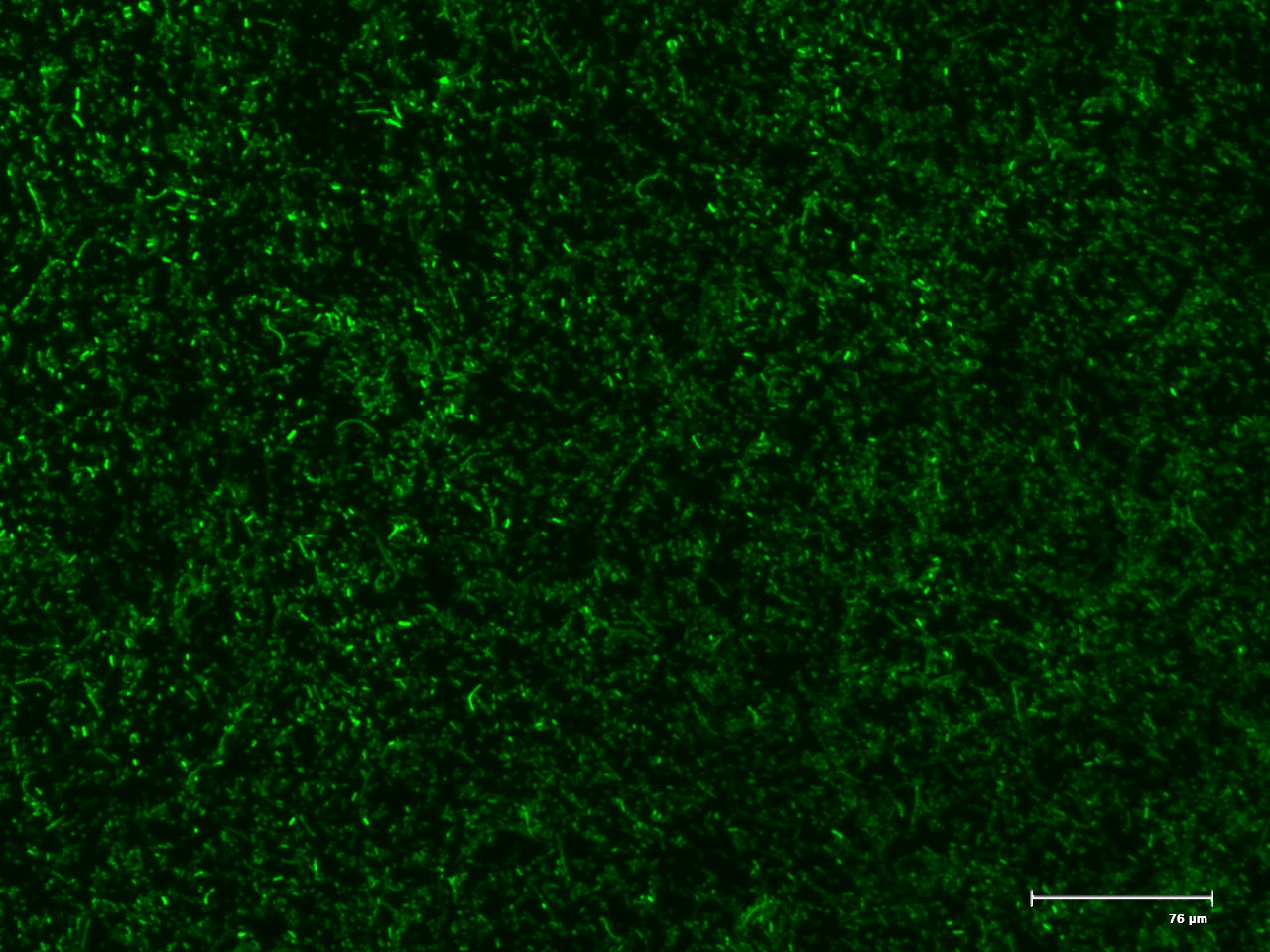
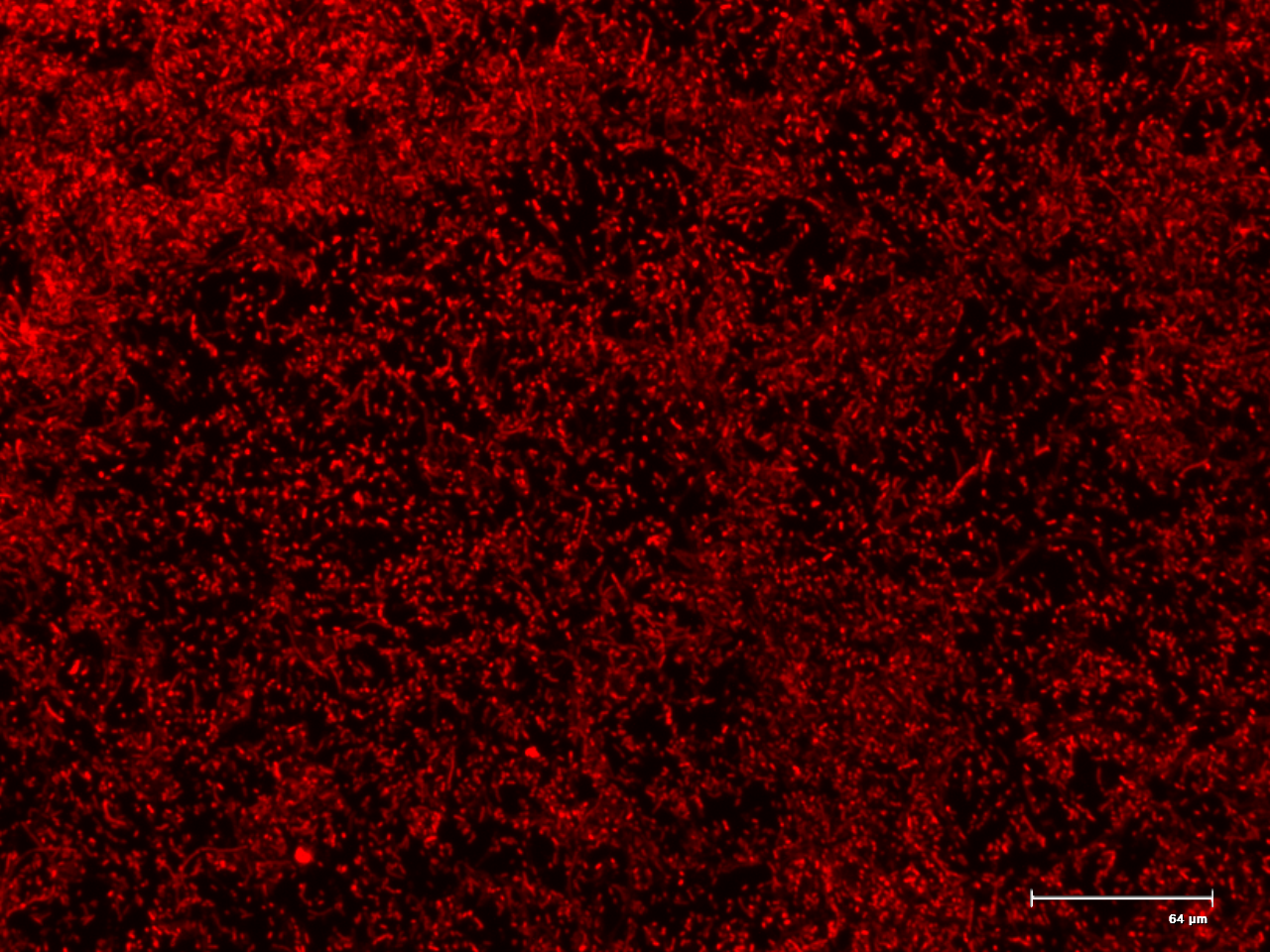
Figure 1. The fluorescent microscopy positive controls GFP (BBa_I13521) and RFP (BBa_I13522).
Cloning of Parts
The parts required for the characterisation of an inverter switch as well as a switch based on excision were synthesised by Integrated DNA technologies as part of their promotion to iGEM teams. These gBlocks were received and blunt end cloned into pUC19 plasmids to create high copy number of our parts. These vectors which were then retrieved by restriction digests of the flanking EcoRI and PstI sites for sticky end cloning into the pSB1C. The digests were loaded onto a 1% agarose gel for electrophoresis, the bands corresponding to the respective parts being characterised were excised and purified using a Zymo Gel purification kit (Inverter 1, BBa_K1768001; Inverter 2 BBa_K176800; Cre generator, BBa_K1768004).

Figure 2. Restriction digest of Inverter 1 (Lane 1), Inverter 2 (Lane 2), Cre generator (Lane 3) and their respective undigested plasmids (Lanes4-6).
The purified inserts were ligated into pSB1C for part submission and the Cre generator BBa_K1768004 ligated into pSB1A for experimentation. These vectors were supplied with the 2015 Distribution Kit and were digested with EcoRI and PstI to create complimentary sticky ends to the inserts. Cultures were grown from the resulting colonies and screened for biobrick parts with the standard VF and VR primer combination. The amplicons were electrophoresed and the size of fragments were referenced to the expected sizes of the fragments for the respective parts.

Figure 3. PCR of biobrick parts in pSB plasmids. Lanes 1,2 Cre generator in pSB1A; Lanes 3,4, Cre generator; Lanes 5,6 Inverter 1; Lanes 7,8 GFP Reverse Compliment, Lanes 9,10 Recombinase ON switch.
<3>Sequencing Parts
The parts were sequenced in the pSB plasmids to confirm their integrity by Sanger method using the sequencing facility at The University of Pretoria. The reactions were set up using BigDye 3.1, its buffer and 150 ng of the respective plasmids as template. Forward and reverse reactions were run using VF and VR primers. Unfortunately when analysing the sequence data it was found that the Cre generator (BBa_K1768004) construct contained two significant mutations in the coding region of the construct. The seven bp deletion causes frame shirt and would render the recombinase protein deactivated.
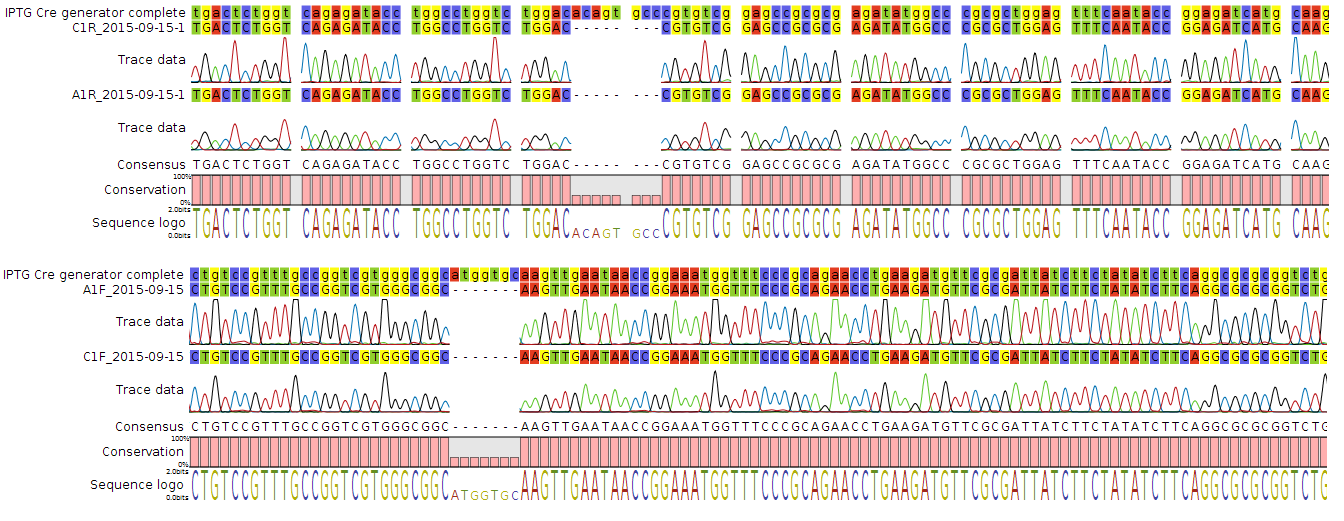
Figure 4. Chromatogram showing the two deletions in the Cre construct from two different sequenced constructs.
Unfortunately due to time constraints the project will have to be completed after the jamboree where the original synthesised gBlock for the Cre recombinase will be re-cloned. Once we have a functional plasmid set we will commence with characterisation of the various switches proposed.


