Team:Pretoria UP/Description
Project Description
Background to the problem
The ability to program the chemotactic behaviour of motile bacteria has piqued the interest of many synthetic biologists, and has important implications for studying cellular communication, environmental applications and human health. In 2011, the WITS-CSIR_SA iGEM team presented a "bio-tweet" concept based on chemotaxis. In their design, motile E. coli use riboswitch-coupled CheZ chemotaxis signaling proteins to travel from a point A to the source of an attractant (theophylline) at point B, then reverse their direction once a concentration threshold has been exceeded to "tweet" back to their starting position, A, through chemotactic motility to a second attractant (atrazine). The team characterized two theophylline riboswitches, but did not characterize their proposed toggle switch.
We recognized that, to resemble a true "bio-tweet", the reply tweet should be both synchronous and conditional on the state of the recipient (B), since reply tweets are never automated responses. The reply tweet should also be able to convey new information about the initial recipient (at point B), for example the state of the local environment. Furthermore, the destination of each "bio-tweet" should ideally be digitally programmed within the cell. The limitations of the "bio-tweet" concept proposed by WITS-CSIR_SA are thus: (1) motile bacteria travel at different rates, and each will reach the critical concentration threshold of an attractant (theophylline) at different times, resulting in unreliable and asynchronous return back to point A; (2) if the bacteria are too sensitive to the theophylline attractant at point B, they will turn around before reaching the first recipient; (3) the "reply tweet" is automatic and the conditions at point B have no bearing on whether or not a reply tweet will be sent, and (4) the information contained in the "bio-tweet" does not change during the reply.
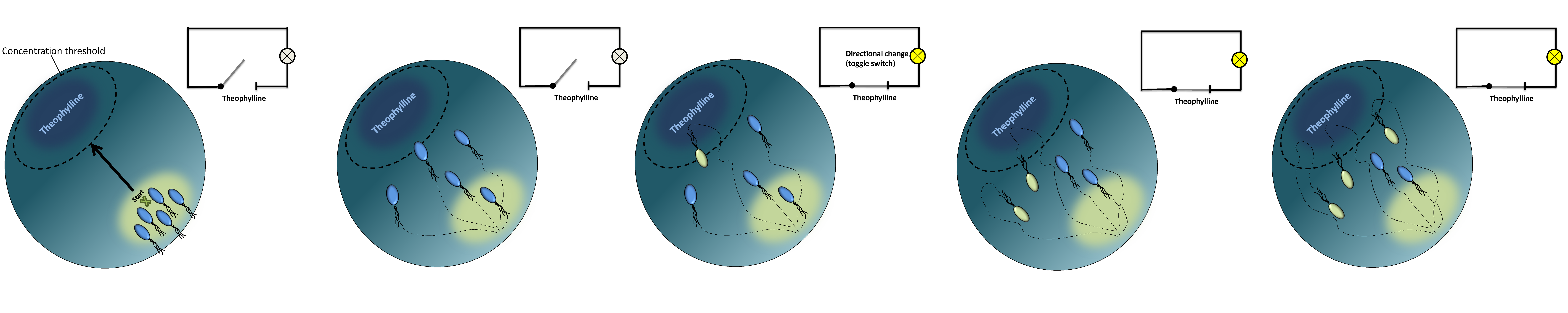
We propose several modifications to the "bio-tweet" principle to satisfy the limitations mentioned above, through five functional modules (Figure 1). A CheZ-coupled riboswitch B programs chemotaxis toward a theophylline attractant, with a lox-based theophylline-induced Cre recombinase toggle switch bringing about chemotaxis reversal as proposed previously. However, we program the induction of Cre to be conditional not only on a theophylline concentration threshold, but also on a quorum sensing module to ensure that a large enough quorum must reach the theophylline threshold before the toggle switch is engaged (BBa_K1768002). An "AND" logic gate is employed to activate the Cre recombinase, relying on input signals from both the quorum sensing module as well as a theophylline-inducible input signal in the form of a protein-protein interaction. This design prevents a quorum signal from inducing Cre expression when the bacteria are at the starting position. Only once the bacteria are at quorum around the theophylline attractant will a transcriptional activator complex induce the promoter upstream of the Cre gene (BBa_K1768004). The Cre recombinase will in turn trigger the inverter switch (BBa_K1768001 or BBa_K1768003), programming the bacteria to synchronously return back to the start. Finally, we include a reporter module that provides information on the state of a chemical of interest, X, at point B. If the chemical is present, an X-inducible promoter is activated and an RFP reporter protein is expressed. The RFP is fused to a transcription factor that self-activates its own promoter, ensuring continued expression of the RFP once the bacteria have moved away from the chemical.


Figure 1. Schematic representation of Switch-coli, an improved bio-tweet design. A theophylline riboswitch (BBa_K249026) activates the ipgC transcription factor (BBa_K1325000) on binding theophylline. A quorum sensing module (BBa_J23119 and BBa_I13263) activates expression of an mxiE chaperone (BBa_K1768002), forming a functional complex that then activates the pipaH promoter (BBa_K1325004) driving Cre recombinase expression. The Cre then engages the toggle switch module using lox-based recombination, reversing the direction of a promoter driving a second, atrazine-activated riboswitch (A) fused to the CheZ chemotaxis-inducing protein.
In our project for 2015, we concentrated on the design and testing of several strategies for the recombinase-based genetic toggle switch module. Transcriptional switches form an integral part of synthetic biology. In the absence of positive feedback loops these systems are unstable since they require the maintained presence of a regulator. DNA switches based on heritable genetic modifications can allow for a permanent, digital change in gene expression even in the absence of the initial signal. The Cre-Lox recombinase system from the P1 bacteriophage has been successfully used in genetic manipulation to excise targeted DNA fragments. The WITS-CSIR_SA team proposed an inversion-based toggle switch, but did not document or submit the corresponding parts or characterise this system. Our aim is to use the Cre-Lox recombinase system to trigger a heritable genetic switch that allows for irreversible ON/OFF programming as an alternative to positive feedback loops. We are testing different approaches relying on the inversion and excision of lox-flanked DNA parts. Our long term goal is to integrate this system with signals from quorum sensing and a logic gate to program conditional, synchronized chemotaxis in motile E. coli.
Inverter Switch Mechanism
An inverter BioBrick (BBa_K1768001 or BBa_K1768003) initially expresses an upstream green fluorescent protein (GFP) marker gene (BBa_K1768000) from a constitutive promoter. This promoter is flanked by two asymmetric Lox elements (Lox66 and Lox71) which are recognised by the Cre recombinase protein. Isopropyl β-D-1-thiogalactopyranoside (IPTG) induction of a pLac promoter results in the expression of Cre which then binds to the Lox elements and mediates recombination between the two sites. This recombination changes the orientation of the Lox-flanked constitutive promoter and results in a mutated Lox site which is not recognised by Cre - preventing further recombination events. The DNA segment between the Lox sites is inverted relative to the flanking DNA and thus the constitutive promoter initiates transcription for the opposite strand and the red fluorescent protein (RFP) marker gene is expressed.
Excision Switch Mechanism
An excision switch BioBrick (BBa_K1768006) initially expresses a green fluorescent protein (GFP) marker gene from a constitutive promoter. The reporter GFP is flanked by two Lox elements, such that the Lox66 site is between the promoter and GFP start codon with the Lox71 downstream of a terminator site that insulates red fluorescent protein (RFP) from transcription. The Lox sites are recognised by the Cre recombinase protein. IPTG induction of a pLac promoter results in the expression of Cre which then binds to the Lox elements and mediates recombination between the two sites. This recombination results in the excision of the GFP gene and terminator. The constitutive promoter is then able to initiate transcription of the RFP reporter gene.
Characterisation
For the two recombinase switches that are being characterised we expect to see a population of green fluorescent E. coli prior to recombination induction. After IPTG-mediated induction of Cre in liquid cultures we expect to observe red fluorescing bacteria in the population. The efficiency of genetic switch function in the bacterial population can be determined by counting the number of RFP-expressing cells relative to the total number of bacteria. We can also assess the fidelity of the genetic switch by sequencing single colonies of RFP-fluorescing cells after plating out the cultures on solid agar plates.
Gilbertson, L. (2003). Cre–lox recombination: Cre-ative tools for plant biotechnology. Trends in biotechnology, 21(12), 550-555.
Ghosh, K., & Van Duyne, G. D. (2002). Cre–loxP biochemistry. Methods, 28(3), 374-383.
Hoess, R. H., & Abremski, K. (1985). Mechanism of strand cleavage and exchange in the Cre-lox site-specific recombination system. Journal of molecular biology, 181(3), 351-362.
Moon, T.S., Lou, C., Tamsir, A., Stanton, B.C. and Voigt, C.A. (2012). Genetic programs constructed from layered logic gates in single cells. Nature, 491(7423): 249–253.
Garg, N., Manchanda, G., & Kumar, A. (2014). Bacterial quorum sensing: circuits and applications. Antonie Van Leeuwenhoek, 105(2), 289-305.




