Difference between revisions of "Team:Goettingen/Results"
| (45 intermediate revisions by 4 users not shown) | |||
| Line 39: | Line 39: | ||
} | } | ||
| − | #menu1, #menu2, #menu3{ | + | #menu1, #menu2, #menu3, #menu4, #menu5, #menu6, #menu7, #menu8{ |
display:none; | display:none; | ||
width:90%; | width:90%; | ||
| Line 62: | Line 62: | ||
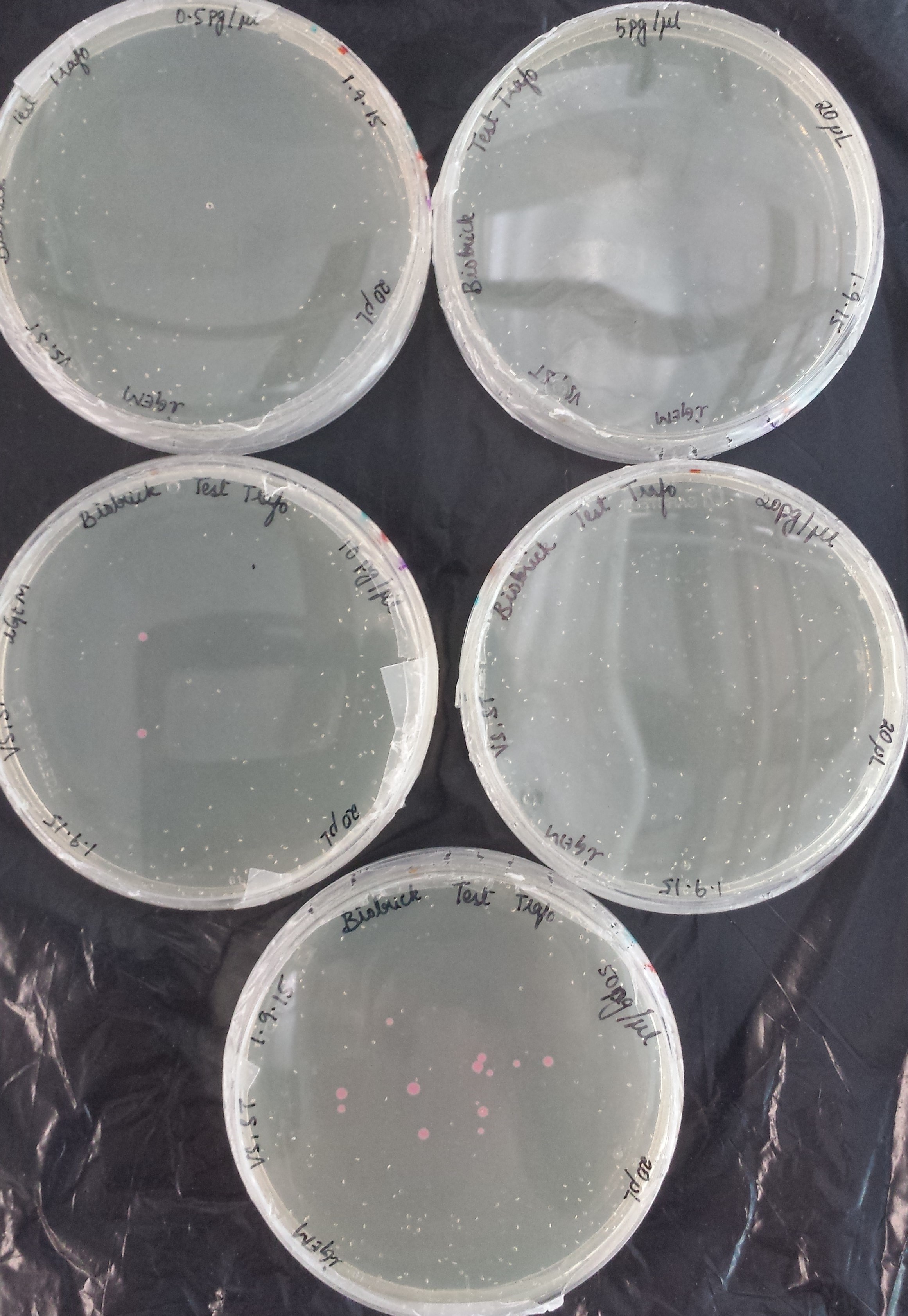
<p>Before using our competent <em>E. coli</em> TOP10 cells in the important experiments, we used the Transformation Efficiency Kit to test the efficiency of our competent cells!</p> | <p>Before using our competent <em>E. coli</em> TOP10 cells in the important experiments, we used the Transformation Efficiency Kit to test the efficiency of our competent cells!</p> | ||
| − | <p>The kit includes five vials of each different DNA concentration: | + | <p>The kit includes five vials of each different DNA concentration: 50 pg/μl, 20 pg/μl, 10 pg/μl, 5 pg/μl, 0.5 pg/μl of purified DNA from BBa_J04450 (RFP construct) in plasmid backbone pSB1C3.</p> |
<p> </p> | <p> </p> | ||
<p>The first test transformation (50 μl of competent cells, 20 μl plated) showed very poor results:</p> | <p>The first test transformation (50 μl of competent cells, 20 μl plated) showed very poor results:</p> | ||
| Line 76: | Line 76: | ||
</td> | </td> | ||
<td> | <td> | ||
| − | <p align="center">0. | + | <p align="center">0.5 pg/μl</p> |
</td> | </td> | ||
<td> | <td> | ||
| − | <p align="center"> | + | <p align="center">5 pg/μl</p> |
</td> | </td> | ||
<td> | <td> | ||
| − | <p align="center"> | + | <p align="center">10 pg/μl</p> |
</td> | </td> | ||
<td> | <td> | ||
| − | <p align="center"> | + | <p align="center">20 pg/μl</p> |
</td> | </td> | ||
<td> | <td> | ||
| − | <p align="center"> | + | <p align="center">50 pg/μl</p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 122: | Line 122: | ||
</td> | </td> | ||
<td> | <td> | ||
| − | <p align="center">3. | + | <p align="center">3.8x 10<sup>6</sup></p> |
</td> | </td> | ||
<td> | <td> | ||
| Line 128: | Line 128: | ||
</td> | </td> | ||
<td> | <td> | ||
| − | <p align="center">3. | + | <p align="center">3.4x 10<sup>6</sup></p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 149: | Line 149: | ||
</td> | </td> | ||
<td valign="bottom" nowrap="nowrap" width="68"> | <td valign="bottom" nowrap="nowrap" width="68"> | ||
| − | <p align="right">1. | + | <p align="right">1.4x 10<sup>6</sup></p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 157: | Line 157: | ||
</td> | </td> | ||
<td valign="bottom" nowrap="nowrap" width="68"> | <td valign="bottom" nowrap="nowrap" width="68"> | ||
| − | <p align="right">3. | + | <p align="right">3.5x 10<sup>6</sup></p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 177: | Line 177: | ||
</td> | </td> | ||
<td> | <td> | ||
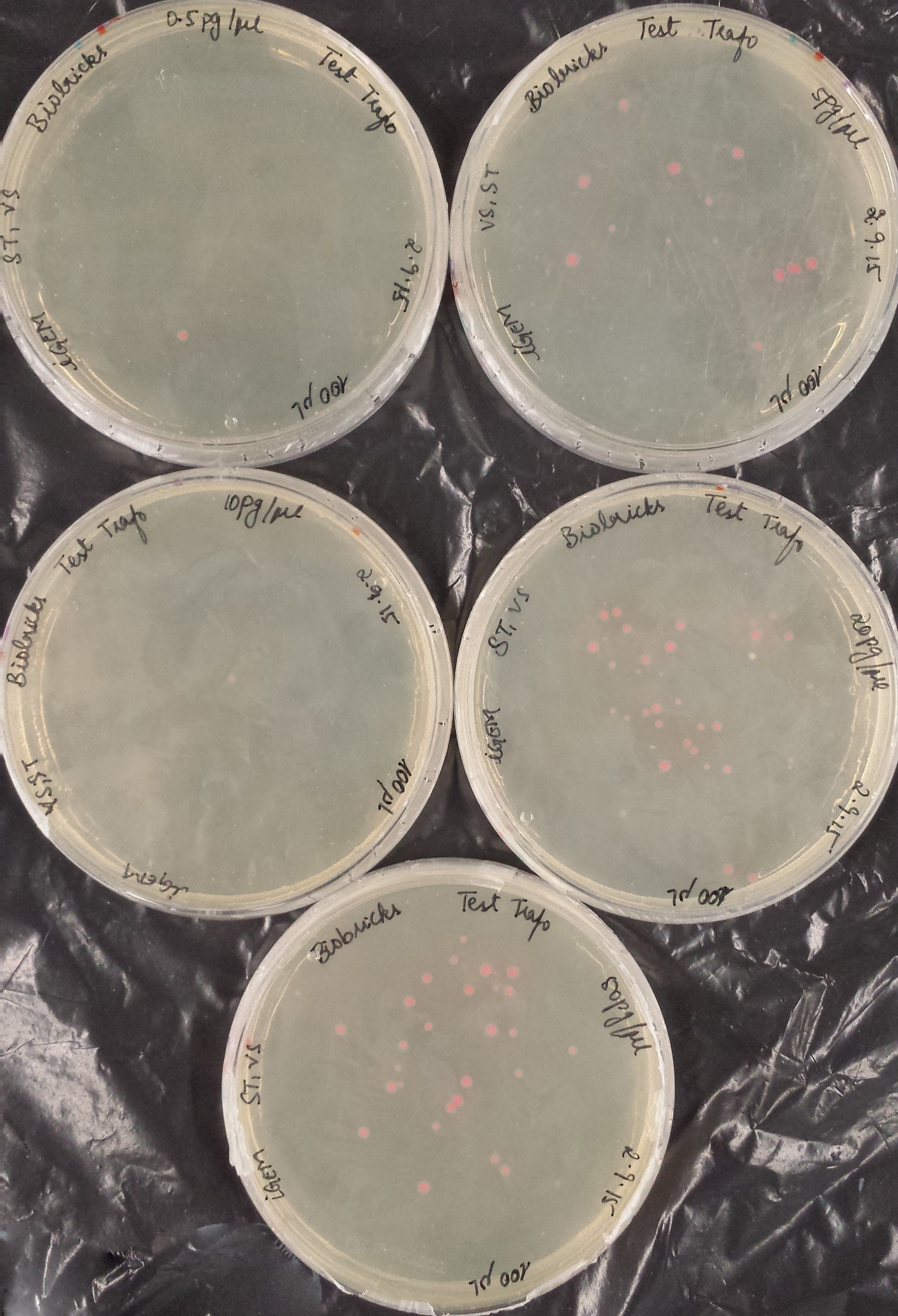
| − | <p align="center">0. | + | <p align="center">0.5 pg/μl</p> |
</td> | </td> | ||
<td> | <td> | ||
| − | <p align="center"> | + | <p align="center">5 pg/μl</p> |
</td> | </td> | ||
<td> | <td> | ||
| − | <p align="center"> | + | <p align="center">10 pg/μl</p> |
</td> | </td> | ||
<td> | <td> | ||
| − | <p align="center"> | + | <p align="center">20 pg/μl</p> |
</td> | </td> | ||
<td> | <td> | ||
| − | <p align="center"> | + | <p align="center">50 pg/μl</p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 217: | Line 217: | ||
</td> | </td> | ||
<td> | <td> | ||
| − | <p align="center">8. | + | <p align="center">8.0x 10<sup>6</sup></p> |
</td> | </td> | ||
<td> | <td> | ||
| − | <p align="center">1. | + | <p align="center">1.0x 10<sup>7</sup></p> |
</td> | </td> | ||
<td> | <td> | ||
| − | <p align="center">4. | + | <p align="center">4.0x 10<sup>5</sup></p> |
</td> | </td> | ||
<td> | <td> | ||
| − | <p align="center">7. | + | <p align="center">7.0x 10<sup>6</sup></p> |
</td> | </td> | ||
<td> | <td> | ||
| − | <p align="center">2. | + | <p align="center">2.3x 10<sup>6</sup></p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 250: | Line 250: | ||
</td> | </td> | ||
<td valign="bottom" nowrap="nowrap" width="68"> | <td valign="bottom" nowrap="nowrap" width="68"> | ||
| − | <p align="right">5. | + | <p align="right">5.6x 10<sup>6</sup></p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 258: | Line 258: | ||
</td> | </td> | ||
<td valign="bottom" nowrap="nowrap" width="68"> | <td valign="bottom" nowrap="nowrap" width="68"> | ||
| − | <p align="right">3. | + | <p align="right">3.7x 10<sup>6</sup></p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 299: | Line 299: | ||
<p> </p> | <p> </p> | ||
<p><strong>MICROSCOPY</strong></p> | <p><strong>MICROSCOPY</strong></p> | ||
| − | <p><span lang="EN-US">To check the activity of RFP, an induction series with 0,2%, 2% and 5% L-Arabinose in the cell culture medium was prepared. Fluorescence microscopy was performed with the RFP DsRed filter (excitation at 536 nm, emission at 582 nm). Unfortunately no fluorescence could be detected throughout the whole project with different constructs. Neither in the pJET_RFP_3, nor in the pBAD constructs (pBAD_RFP_3 and pBAD_RFP_ACEL) induced with 0,2%, 2% and 5% L-Arabinose in the medium. | + | <p><span lang="EN-US">To check the activity of RFP, an induction series with 0,2%, 2% and 5% L-Arabinose in the cell culture medium was prepared. Fluorescence microscopy was performed with the RFP DsRed filter (excitation at 536 nm, emission at 582 nm). Unfortunately no fluorescence could be detected throughout the whole project with different constructs. Neither in the pJET_RFP_3, nor in the pBAD constructs (pBAD_RFP_3 and pBAD_RFP_ACEL) induced with 0,2%, 2% and 5% L-Arabinose in the medium. Since the DNA sequences have been confirmed to be correct by Sanger sequencing it might be a problem of expression. |
<p> </p> | <p> </p> | ||
<p><strong>FUTURE PLANS</strong></p>In future we want to exchange our vector system (pET100 instead of pBAD) to ensure that pBAD is not somehow interfering with our chosen <em>E. coli</em> strain.</span></p> | <p><strong>FUTURE PLANS</strong></p>In future we want to exchange our vector system (pET100 instead of pBAD) to ensure that pBAD is not somehow interfering with our chosen <em>E. coli</em> strain.</span></p> | ||
| Line 312: | Line 312: | ||
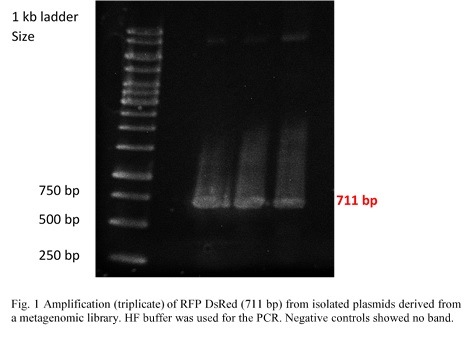
<p> | <p> | ||
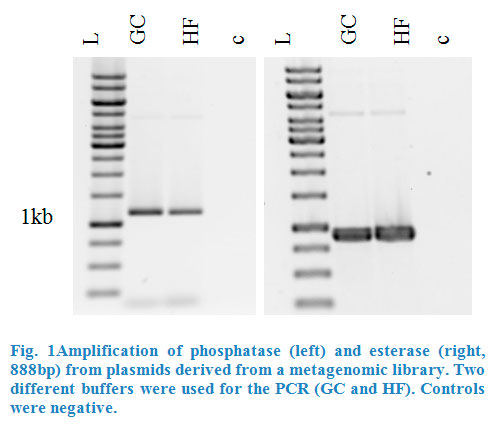
Esterase was amplified from pET101_E064 and phosphatase from pTOPOXL_PLP07 by PCR for each enzyme (Fig.1). Both plasmids were given to us by the Applied | Esterase was amplified from pET101_E064 and phosphatase from pTOPOXL_PLP07 by PCR for each enzyme (Fig.1). Both plasmids were given to us by the Applied | ||
| − | and Genomic Microbiology department. Primers contained restriction sites | + | and Genomic Microbiology department. Primers contained restriction sites <em>Kpn</em>I and <em>Sac</em>I for esterase and <em>Xho</em>I and <em>Pst</em>I for the phosphatase in order to make |
| − | them compatible for insertion into the multiple cloning site of the pBAD vector .The genes for these two enzymes had been found in screenings of | + | them compatible for insertion into the multiple cloning site of the pBAD vector. The genes for these two enzymes had been found in screenings of |
metagenomic libraries. | metagenomic libraries. | ||
</p> | </p> | ||
| Line 326: | Line 326: | ||
<p> | <p> | ||
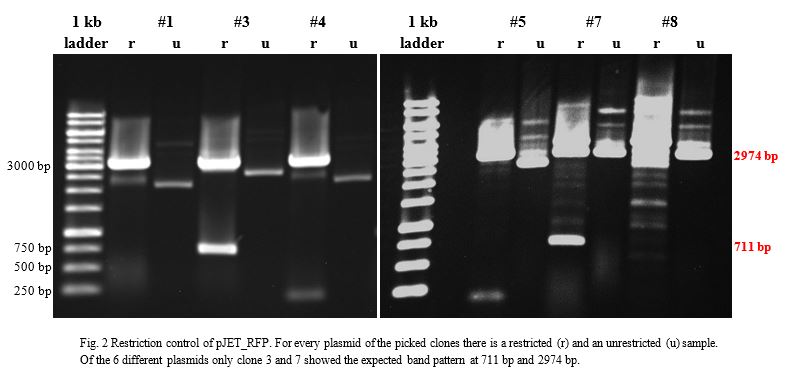
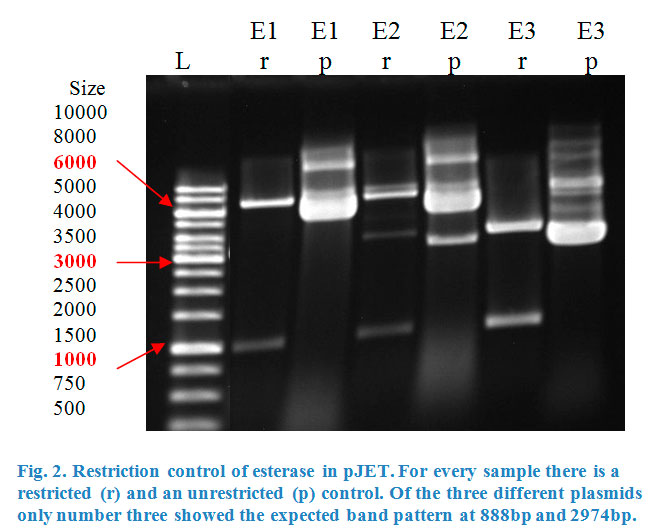
Ligation into pJET was followed according to the protocol in the methods collection. After over-night incubation colonies were picked, plasmids extracted | Ligation into pJET was followed according to the protocol in the methods collection. After over-night incubation colonies were picked, plasmids extracted | ||
| − | with a Quiaprep spin Miniprep kit and restricted with either | + | with a Quiaprep spin Miniprep kit and restricted with either <em>Kpn</em>I and <em>Sac</em>I or <em>Xho</em>I and <em>Pst</em>I, depending on the Enzyme (Fig.2&3). |
</p> | </p> | ||
</html> | </html> | ||
| Line 347: | Line 347: | ||
</p> | </p> | ||
<p> | <p> | ||
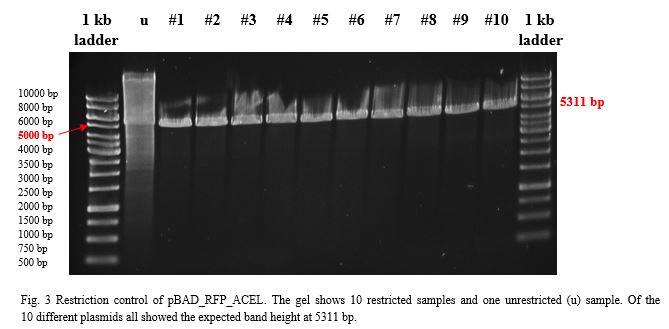
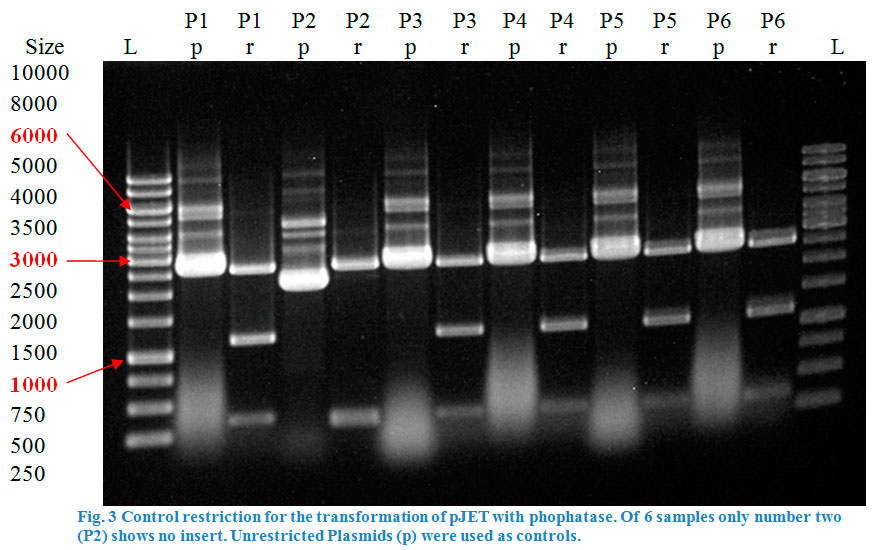
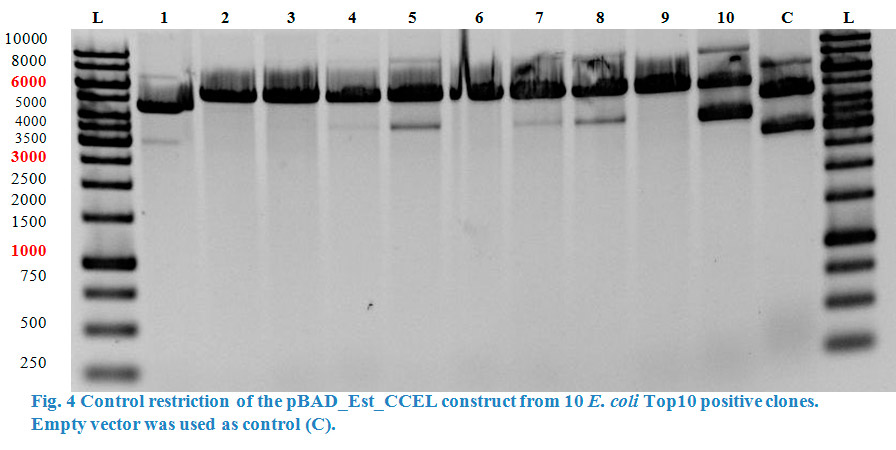
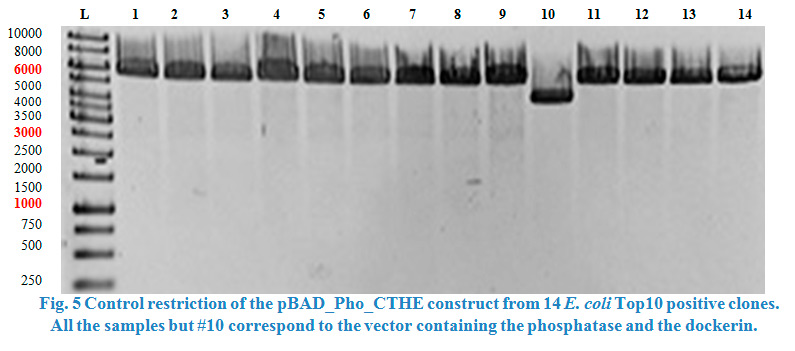
| − | A restriction control with | + | A restriction control with <em>Sac</em>I for the esterase constructs was run in a 0,8% agarose gel (Fig. 4). Samples #2, #3 and #9 correspond<strong> </strong>to the linear plasmid containing the insert. Phosphatase construct was control restricted with <em>EcoR</em>I (Fig. 5), in this case 13 of 14 samples showed successful insertion of the enzyme fragment into the vector. |
| − | + | ||
</p> | </p> | ||
</html> | </html> | ||
| − | [[File: | + | [[File:Control restriction pBAD Est CCEL iGEM Goettingen2015.jpeg|600px|thumb|center|]] |
<html> | <html> | ||
| + | </html> | ||
| + | [[File:Pho_CTHE_control_restriction_iGEM_Gottingen2015.jpg |600px|thumb|center|]] | ||
| + | <html> | ||
| + | |||
<p> | <p> | ||
| − | + | <strong>Future plans</strong> | |
</p> | </p> | ||
| + | <p> | ||
| + | The next steps include changing to a better expression vector in order to test and measure the activity of the enzymes, especially to ensure that the fusion with the dockerins is not interfering with the function of the enzymes. Finally the aim is to combine the purified enzyme-dockerin proteins with the synthetic scaffold and prove the self assemby of the Flexosome tool. | ||
| + | </p> | ||
| + | </div> | ||
| − | + | <a href="" onClick=" $('#menu5').slideToggle(300, function callback() { }); return false;"><h1>Cellulase</h1></a> | |
| + | <div id="menu5"><p> | ||
| + | |||
| + | </p> | ||
| + | <p> | ||
| + | The cellulase gene was amplified using PCR. The primers which were used contained the restriction sites for ligation into the pBAD vector. The cellulase | ||
| + | PCR products were then checked on 0.8 % agarose gel. | ||
| + | </p> | ||
| + | </html> | ||
| + | [[File:TeamGoettingen_CellulasePCR.jpg|300px|center|thumb|Figure 1: Cellulase PCR product was run on a 0.8 % agarose gel. The desired cellulase sequence is at 2496 bp.]] | ||
| + | <html> | ||
| + | <p> | ||
| + | The cellulase PCR products were then purified by using the <strong>QIAquick® PCR Purification Kit (QIAGEN). </strong>After the PCR products were purified, | ||
| + | the concentration of DNA was measured by Nanodrop 2000c Spectrophotometer.<strong></strong> | ||
| + | </p> | ||
| + | <p> | ||
| + | <strong>Restriction control of pBAD_CCEL and cellulase PCR product</strong> | ||
| + | </p> | ||
| + | <p> | ||
| + | The purified cellulase PCR product and the pBAD_CCEL vector were restricted by following a double digest protocol with SacI and PvuII enzymes. After | ||
| + | restriction control products were purified by using the <strong>QIAquick® PCR Purification Kit (QIAGEN) </strong>the concentration of DNA was measured by | ||
| + | Nanodrop 2000c Spectrophotometer.<strong></strong> | ||
| + | </p> | ||
| + | <p> | ||
| + | <strong>Ligation of Cellulase into pBAD- CCEL</strong> | ||
| + | </p> | ||
| + | <p> | ||
| + | The cellulase gene was then ligated into the pBAD vector containing the CCEL (<em>Clostridium</em> | ||
| + | </p> | ||
| + | <p> | ||
| + | <em>cellulolyticum</em> | ||
| + | ) dockerin. | ||
| + | </p> | ||
| + | <p> | ||
| + | For ligation of the cellulase PCR product into the restricted pBAD_CCEL vector, the Thermo Fisher T4 Ligase Sticky End Ligation protocol was followed. | ||
| + | </p> | ||
| + | <p> | ||
| + | <strong>Transformation of pBAD_CellulaseCCEL into chemically competent Top10 <em>E. coli </em>cells</strong> | ||
| + | </p> | ||
| + | <p> | ||
| + | pBAD_CellulaseCCEL was transformed into competent Top10 <em>E. coli </em>cells by using the heat shock transformation method. | ||
| + | </p> | ||
| + | </html> | ||
| + | [[File:TeamGoettingen_pBAD_CellulaseCCEL_Trafo.jpg|300px|center|thumb|Figure 2: Transformation of pBAD_CellulaseCCEL into competent Top 10 E. coli cells. The image shows plates with successful transformants.]] | ||
| + | <html> | ||
| + | <p> | ||
| + | Colonies from transformation plates were inoculated into LB medium containing ampicillin (100 µg/ml). | ||
| + | </p> | ||
| + | <p> | ||
| + | <strong>Plasmid Extraction: peqGold Plasmid Miniprep Kit</strong> | ||
| + | </p> | ||
| + | <p> | ||
| + | Plasmid extraction was done for colonies inoculated in LB medium by using peqGold Plasmid Miniprep kit and plasmid concentration was measured by using | ||
| + | Nanodrop 2000c Spectrophotometer. | ||
| + | </p> | ||
| + | <p> | ||
| + | Restriction control was done with the enzymes SacI and PvuII from Thermo fisher scientific.<strong> </strong>Restricted plasmids were checked on a 0.8 % | ||
| + | agarose gel. | ||
| + | </p> | ||
| + | </html> | ||
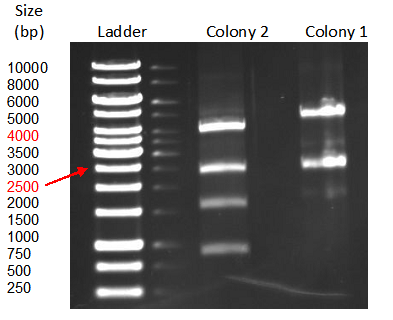
| + | [[File:TeamGoettingen_Restriction_pBAD_CellulaseCCEL.jpg|300px|center|thumb|Figure 3: Restriction control of the pBAD_CellulaseCCEL construct from TOP10 E. coli positive clones by with SacI and PvuII. Colony 1 has the desired insert.]] | ||
| + | <html> | ||
| + | <p> | ||
| + | The correct insert was confirmed through sequencing. | ||
| + | </p> | ||
</div> | </div> | ||
| + | <a href="" onClick=" $('#menu4').slideToggle(300, function callback() { }); return false;"><h1>BioBricks</h1></a> | ||
| + | <div id="menu4"> | ||
| − | < | + | <p> |
| − | + | <strong>Dockerin Biobricks</strong> | |
| − | <p> | + | </p> |
| − | < | + | <p> |
| − | < | + | The dockerins ACEL (<em>Acetivibrio cellulolyticus</em>), BCEL (<em>Bacteroides cellulosolvens</em>), CCEL (<em>Clostridium cellulolyticum</em>) and CTHE (<em>Clostridium thermocellum</em>) were amplified from pBAD_ACEL, pBAD_BCEL, pBAD_CCEL and pBAD_CTHE with primers to add the desired <em>Eco</em>RI and <em>Pst</em>I restriction sites by PCR. Primers contained restriction sites for <em>Eco</em>RI and <em>Pst</em>I in order to make them compatible for |
| − | < | + | insertion into the iGEM shipping vector pSB1C3. |
| − | < | + | </p> |
| − | </ | + | <p> |
| + | After purification, the PCR products were restricted with <em>Eco</em>RI and <em>Pst</em>I, as well as pSB1C3. Afterwards the restricted dockerins were | ||
| + | ligated into pSB1C3 by T4 ligation (sticky end ligation) and transformed into <em>E. coli</em> TOP10. | ||
| + | </p> | ||
| + | <p> | ||
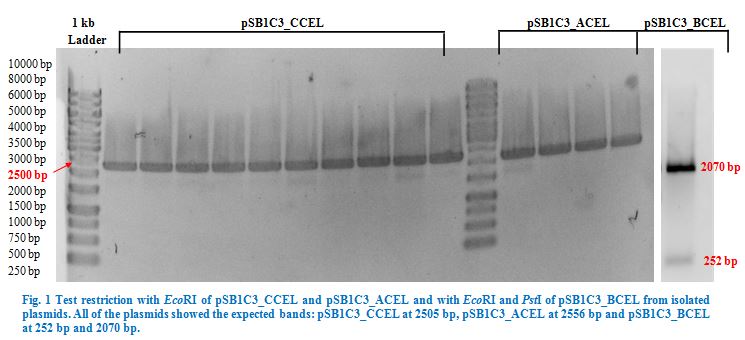
| + | Ligation into pSB1C3 was followed according to the protocol in the methods collection. After over-night incubation colonies were picked, plasmids extracted | ||
| + | with the QIAGEN QIAprep Spin Miniprep Kit and test restricted with <em>Eco</em>RI (pSB1C3_CCEL and pSB1C3_ACEL) and <em>Eco</em>RI and <em>Pst</em>I | ||
| + | (pSB1C3_BCEL) (Fig.1). We lost CTHE due to failing transformations at this point. | ||
| + | </p> | ||
| − | < | + | </html> |
| + | [[File:Rest_control_pSB1C3_ACEL_CCEL_BCEL.jpeg|600px|thumb|center|]] | ||
| + | <html> | ||
| − | < | + | <p> |
| − | + | Once restriction controls showed the correct bands, pSB1C3_CCEL, pSB1C3_ACEL and pSB1C3_BCEL were sent for sequencing. | |
| − | < | + | </p> |
| − | < | + | <p> </p> |
| + | <p> | ||
| + | <strong>Colour BioBricks</strong> | ||
| + | </p> | ||
| + | <p> | ||
| + | We also wanted to improve already existing BioBricks by fusing our dockerins to the colours eforRed (<a href="http://parts.igem.org/Part:BBa_K592012"style="color: #000066">BBa_K592012</a>) and amilCP (<a href="http://parts.igem.org/Part:BBa_K592009"style="color: #000066">BBa_K592009</a>) that were | ||
| + | submitted from the University of Uppsala (Sweden) in 2011. | ||
| + | </p> | ||
| + | <p> | ||
| + | So we chose to fuse eforRed to BCEL and amilCP to CCEL. Since the plasmids of the two colour proteins were not distributed with the current plate of the | ||
| + | iGEM Distribution Kit, we ordered those parts including the right restriction sites as gBlocks from IDT. | ||
| + | </p> | ||
| + | <p> | ||
| + | As a first step eforRed and amilCP were restricted with <em>Kpn</em>I and <em>Sac</em>I to make them compatible with our dockerins in pBAD. | ||
| + | </p> | ||
| + | <p> | ||
| + | After purification restricted colours were ligated into pBAD_BCEL and pBAD_CCEL by T4 ligation (sticky end ligation) and transformed into <em>E. coli</em> | ||
| + | TOP10. | ||
| + | </p> | ||
| + | <p> | ||
| + | Ligation into pBAD was followed according to the protocol in the methods collection. After over-night incubation colonies were picked, plasmids extracted | ||
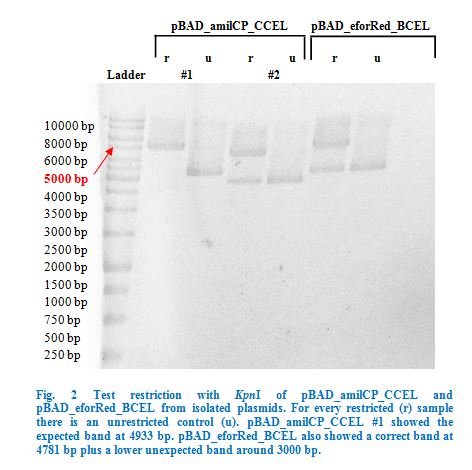
| + | with the QIAGEN QIAprep Spin Miniprep Kit and test restricted with <em>Kpn</em>I (Fig.2). | ||
| + | </p> | ||
| + | </html> | ||
| + | [[File:TeamGoettingen2015_Rest_control_pBAD_eforRed_BCEL_and_amilCP_CCEL_Goettingen2015.jpeg|600px|thumb|center|]] | ||
| + | <html> | ||
| − | < | + | <p> |
| − | <p> | + | Once restriction controls showed the correct bands, pBAD_amilCP_CCEL #1 and pBAD_eforRed_BCEL were sent for sequencing. Sequencing showed that both |
| − | < | + | plasmids were correct. |
| − | < | + | </p> |
| − | < | + | <p> |
| − | < | + | To make our constructs compatible with the iGEM shipping vector pSB1C3 the desired <em>Eco</em>RI and <em>Pst</em>I restrictions sites were added by PCR. |
| − | </ | + | </p> |
| + | <p> | ||
| + | After purification, the PCR products were restricted with <em>Eco</em>RI and <em>Pst</em>I, the same with pSB1C3, and afterwards ligated into pSB1C3 by T4 | ||
| + | ligation (sticky end ligation) and transformed into <em>E. coli</em> TOP10. | ||
| + | </p> | ||
| + | <p> | ||
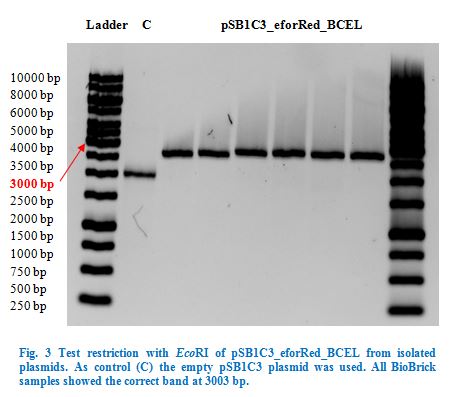
| + | After over-night incubation colonies were picked, plasmids extracted with the QIAGEN QIAprep Spin Miniprep Kit and test restricted with <em>Eco</em>RI | ||
| + | (Fig.3). Due to the reason that our dockerin CCEL contains an internal <em>Pst</em>I restriction site we continued working only with the eforRed_BCEL | ||
| + | construct. | ||
| + | </p> | ||
| + | </html> | ||
| + | [[File:TeamGoettingen2015_Rest_control_pSB1C3_eforRed_BCEL_Goettingen2015.jpeg|600px|thumb|center|]] | ||
| + | <html> | ||
| + | |||
| + | <p> | ||
| + | Once restriction controls showed the correct band, pSB1C3_eforRed_BCEL was sent for sequencing. Sequencing showed that the plasmids were correct. | ||
| + | </p> | ||
| + | <p> | ||
| + | <strong>RESULTS</strong> | ||
| + | </p> | ||
| + | <p> | ||
| + | Sequencing showed that the dockerins ACEL and CCEL contain an internal <em>Pst</em>I restriction site. Therefore all the constructs containing ACEL or CCEL | ||
| + | showed in the end truncated dockerin sequences and could not be send in as BioBricks. | ||
| + | </p> | ||
| + | <p> | ||
| + | We also build a construct where the enzyme esterase was fused to our CCEL dockerin (pBAD_Est_CCEL) following the same strategy. But again, due to the | ||
| + | reason that our dockerin CCEL contains an internal <em>Pst</em>I restriction site we had to stop our work at this point. | ||
| + | </p> | ||
| + | <p> | ||
| + | Furthermore we <a href="https://2015.igem.org/Team:Goettingen/Collaborations"style="color: #000066">collaborated</a> with the current iGEM team of Aachen and tried to fuse three of their enzymes to our constructs but could not finish our work here. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | <strong>ACHIEVENTS<a name="_GoBack"></a></strong> | ||
| + | </p> | ||
| + | <p> | ||
| + | <strong>Sequences of pSB1C3_BCEL and pSB1C3_eforRed_BCEL were correct and submitted as BioBricks to iGEM:</strong> | ||
| + | </p> | ||
| + | <p> | ||
| + | pSB1C3_BCEL (<a href="http://parts.igem.org/Part:BBa_K1865000"style="color: #000066">BBa_K1865000</a>) | ||
| + | </p> | ||
| + | <p> | ||
| + | pSB1C3_eforRed_BCEL (<a href="http://parts.igem.org/Part:BBa_K1865001"style="color: #000066">BBa_K1865001</a>) | ||
| + | </p> | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <a href="" onClick=" $('#menu6').slideToggle(300, function callback() { }); return false;"><h1>Scaffoldin Purification</h1></a> | ||
| + | <div id="menu6"> | ||
| + | <p> | ||
| + | <strong>Scaffoldin purification</strong> | ||
| + | </p> | ||
| + | <p> | ||
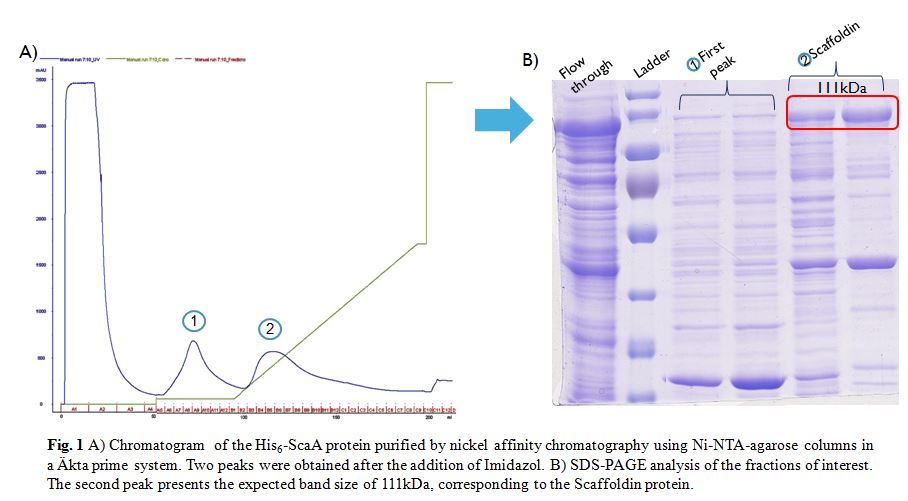
| + | After induction according to the methods collection, the targeted ScaA scaffoldin was purified by nickel affinity chromatography using Ni-NTA-agarose columns in a Äkta prime system (Fig. 1A). Following the addition of Imidazol two peaks were obtained. The fractions of interest were visualized in a SDS-PAGE gel (Fig. 1B). The gel showed that only the second fraction presents the expected size of 111kDa. However some other bands appeared along this fraction, therefore in order to obtain even more pure protein a Size Exclusion Chromatography (SEC) was done. | ||
| + | </p> | ||
| + | </html> | ||
| + | [[File:Histag_purification_iGEM_Goettingen2015.jpeg|600px|thumb|center|]] | ||
| + | <html> | ||
| + | <p> | ||
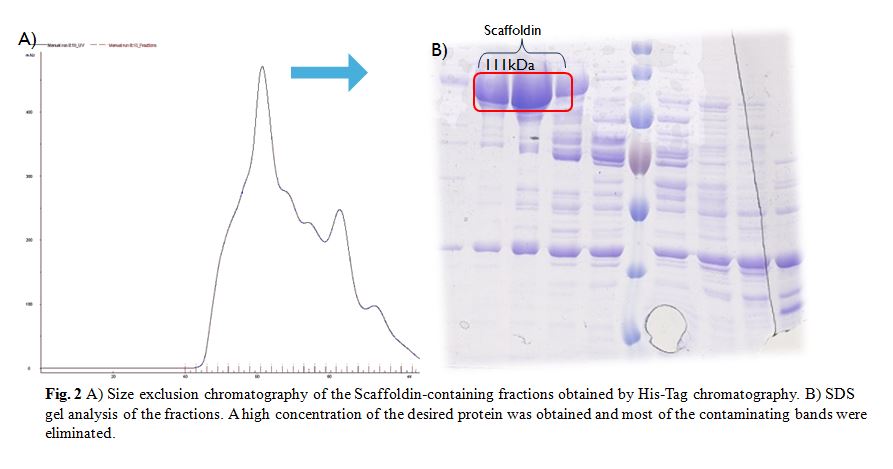
| + | The SEC was done with a HiLoad Superdex 200 16/600 column (GE Healthcare) (Fig. 2A). SDS gel shows that a high concentration of the desired protein was | ||
| + | obtained and most of the previous contaminating bands were eliminated. Nevertheless there was an unknown band of low weight present in every fraction, | ||
| + | apparently it could be a second protein from E. coli with some affinity to the Scaffoldin than is then is co-purified with it (Fig. 2B). | ||
| + | </p> | ||
| + | </html> | ||
| + | [[File:SEC_iGEM_Gottingen2015.jpeg|600px|thumb|center|]] | ||
| + | <html> | ||
| + | <p> | ||
| + | Future plans include to incubate the Scafoldin with the purified enzyme-dockerin construct in order to prove that the Flexosome can assembly by itself. | ||
| + | </p> | ||
| + | </div> | ||
| + | <a href="" onClick=" $('#menu7').slideToggle(300, function callback() { }); return false;"><h1>Copurification</h1></a> | ||
| + | <div id="menu7"> | ||
| + | <p> | ||
| + | Copurification of Scaffoldin and Enzyme-Dockerin | ||
| + | </p> | ||
| + | <p> | ||
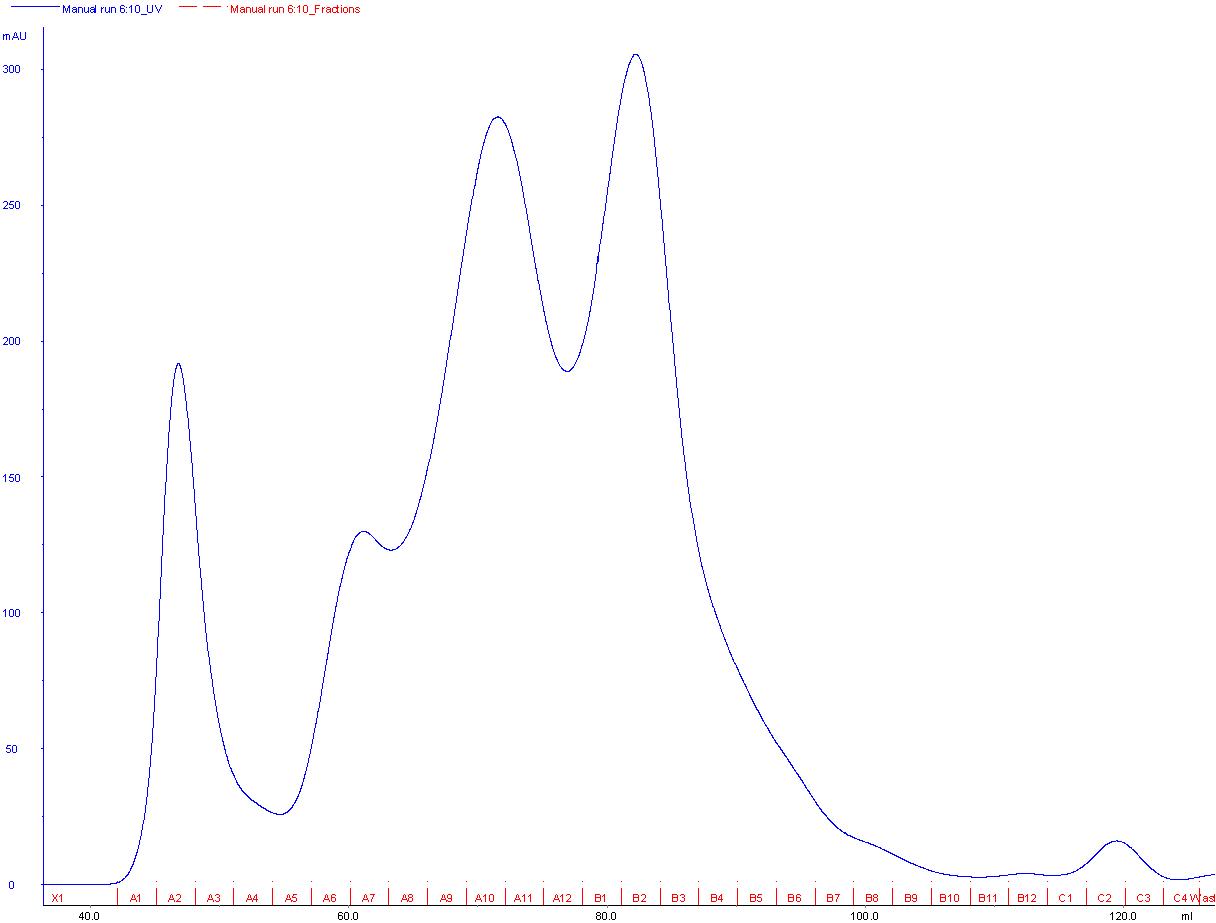
| + | The main peaks after gelfiltration from the scaffoldin CTHE (see results scaffoldin) and CellulaseCCEL (Fig.1) were pooled and concentrated. | ||
| + | </p> | ||
| + | </html> | ||
| + | [[File:TeamGoettingen_GeFi_CellulaseCCEL.jpg|650px|centre|thumb|B1-B3 were pooled for copurification.]] | ||
| + | <html> | ||
| + | <p> | ||
| + | They were then mixed and incubated together at 37°C for 15 min with agitation. | ||
| + | </p> | ||
| + | <p> | ||
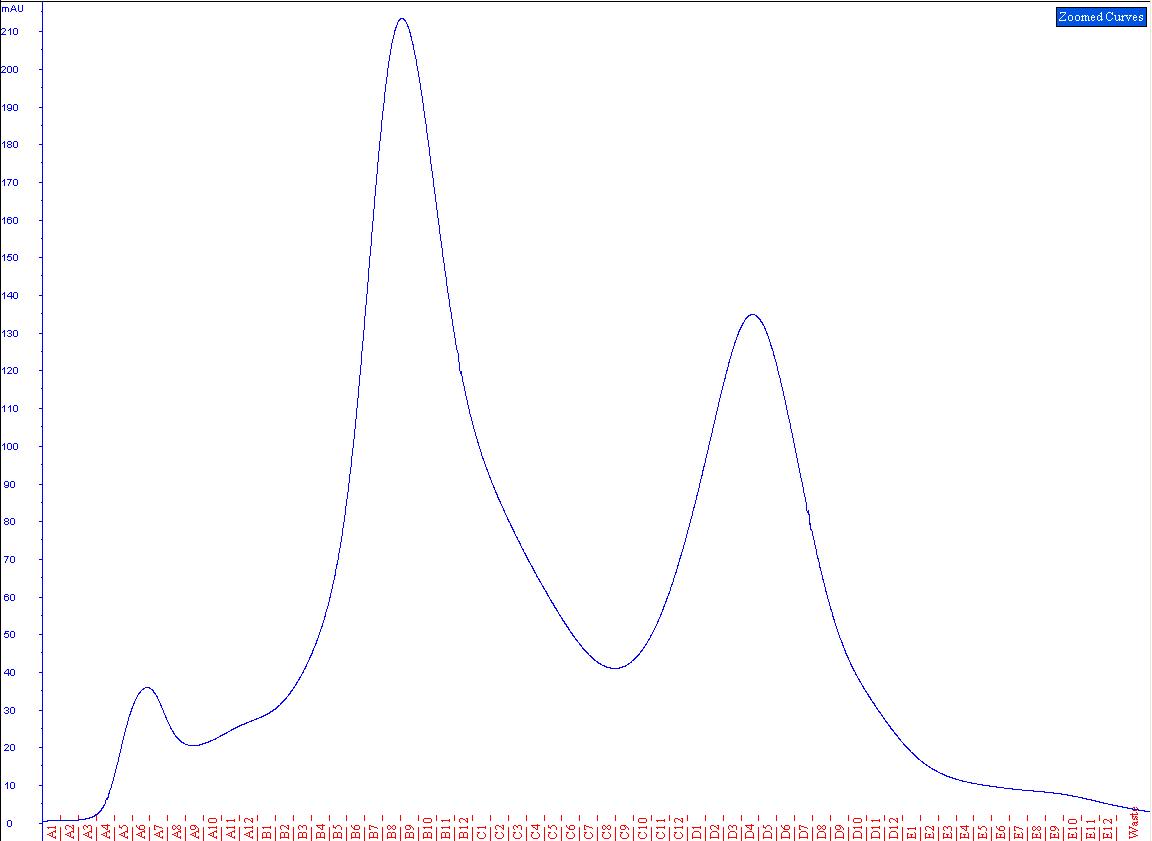
| + | The solution was then used in another gelfiltration (Fig.2). The resulting peaks were run on a SDS-gel (Fig.3) and checked for cellulase activity. | ||
| + | </p> | ||
| + | </html> | ||
| + | [[File:TeamGoettingen_Copurification_CTHE_CellulaseCCEL.jpg|650px|centre|thumb|Fig.2) The peak at A5-A7 may be the bound scaffoldin and dockerin, whilst the unbound proteins are expected between B3 and C9.]] | ||
| + | |||
| + | <html> | ||
| + | <p> | ||
| + | The first peak from the copurification showed a distinct cellulase activity. As the peak ran considerably higher than expected than either the scaffoldin | ||
| + | or the CellulaseCCEL in the gelfitration, it can be expected that this peak contains a successfully formed complex between CellulaseCCEL and the CTHE | ||
| + | scaffoldin. | ||
| + | </p> | ||
| + | </div> | ||
| + | <a href="" onClick=" $('#menu8').slideToggle(300, function callback() { }); return false;"><h1>Giant Jamboree: Poster and Presentation</h1></a> | ||
| + | <div id="menu8"> | ||
| + | <p> | ||
| + | Did you missed our presentation? Do you want to see our poster again? | ||
| + | No problem, here you can find the files: | ||
| + | </p> | ||
| + | <p> | ||
| + | </html> | ||
| + | [[Media:IGEM-presentation_final_team_Goettingen.ppt]] | ||
| + | <html> | ||
| + | </p> | ||
| + | <p> | ||
| + | </html> | ||
| + | [[Media:Poster_teamGoettingen_2015.pdf]] | ||
| + | <html> | ||
| + | </p> | ||
</div> | </div> | ||
</html> | </html> | ||
Latest revision as of 22:03, 20 October 2015