Difference between revisions of "Team:Goettingen/Results"
| (34 intermediate revisions by 3 users not shown) | |||
| Line 39: | Line 39: | ||
} | } | ||
| − | #menu1, #menu2, #menu3, #menu4{ | + | #menu1, #menu2, #menu3, #menu4, #menu5, #menu6, #menu7, #menu8{ |
display:none; | display:none; | ||
width:90%; | width:90%; | ||
| Line 364: | Line 364: | ||
</div> | </div> | ||
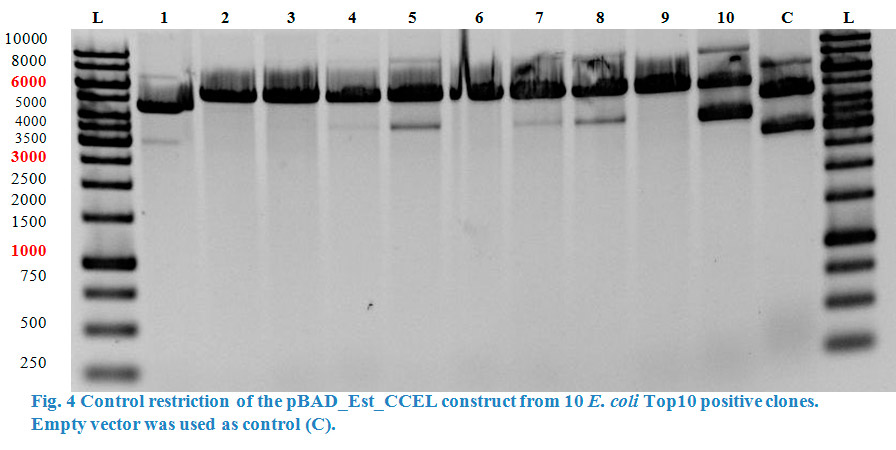
| + | <a href="" onClick=" $('#menu5').slideToggle(300, function callback() { }); return false;"><h1>Cellulase</h1></a> | ||
| + | <div id="menu5"><p> | ||
| + | |||
| + | </p> | ||
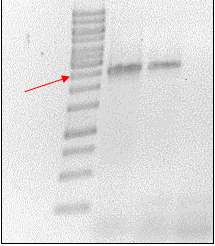
| + | <p> | ||
| + | The cellulase gene was amplified using PCR. The primers which were used contained the restriction sites for ligation into the pBAD vector. The cellulase | ||
| + | PCR products were then checked on 0.8 % agarose gel. | ||
| + | </p> | ||
| + | </html> | ||
| + | [[File:TeamGoettingen_CellulasePCR.jpg|300px|center|thumb|Figure 1: Cellulase PCR product was run on a 0.8 % agarose gel. The desired cellulase sequence is at 2496 bp.]] | ||
| + | <html> | ||
| + | <p> | ||
| + | The cellulase PCR products were then purified by using the <strong>QIAquick® PCR Purification Kit (QIAGEN). </strong>After the PCR products were purified, | ||
| + | the concentration of DNA was measured by Nanodrop 2000c Spectrophotometer.<strong></strong> | ||
| + | </p> | ||
| + | <p> | ||
| + | <strong>Restriction control of pBAD_CCEL and cellulase PCR product</strong> | ||
| + | </p> | ||
| + | <p> | ||
| + | The purified cellulase PCR product and the pBAD_CCEL vector were restricted by following a double digest protocol with SacI and PvuII enzymes. After | ||
| + | restriction control products were purified by using the <strong>QIAquick® PCR Purification Kit (QIAGEN) </strong>the concentration of DNA was measured by | ||
| + | Nanodrop 2000c Spectrophotometer.<strong></strong> | ||
| + | </p> | ||
| + | <p> | ||
| + | <strong>Ligation of Cellulase into pBAD- CCEL</strong> | ||
| + | </p> | ||
| + | <p> | ||
| + | The cellulase gene was then ligated into the pBAD vector containing the CCEL (<em>Clostridium</em> | ||
| + | </p> | ||
| + | <p> | ||
| + | <em>cellulolyticum</em> | ||
| + | ) dockerin. | ||
| + | </p> | ||
| + | <p> | ||
| + | For ligation of the cellulase PCR product into the restricted pBAD_CCEL vector, the Thermo Fisher T4 Ligase Sticky End Ligation protocol was followed. | ||
| + | </p> | ||
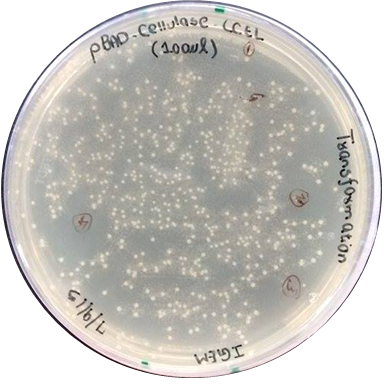
| + | <p> | ||
| + | <strong>Transformation of pBAD_CellulaseCCEL into chemically competent Top10 <em>E. coli </em>cells</strong> | ||
| + | </p> | ||
| + | <p> | ||
| + | pBAD_CellulaseCCEL was transformed into competent Top10 <em>E. coli </em>cells by using the heat shock transformation method. | ||
| + | </p> | ||
| + | </html> | ||
| + | [[File:TeamGoettingen_pBAD_CellulaseCCEL_Trafo.jpg|300px|center|thumb|Figure 2: Transformation of pBAD_CellulaseCCEL into competent Top 10 E. coli cells. The image shows plates with successful transformants.]] | ||
| + | <html> | ||
| + | <p> | ||
| + | Colonies from transformation plates were inoculated into LB medium containing ampicillin (100 µg/ml). | ||
| + | </p> | ||
| + | <p> | ||
| + | <strong>Plasmid Extraction: peqGold Plasmid Miniprep Kit</strong> | ||
| + | </p> | ||
| + | <p> | ||
| + | Plasmid extraction was done for colonies inoculated in LB medium by using peqGold Plasmid Miniprep kit and plasmid concentration was measured by using | ||
| + | Nanodrop 2000c Spectrophotometer. | ||
| + | </p> | ||
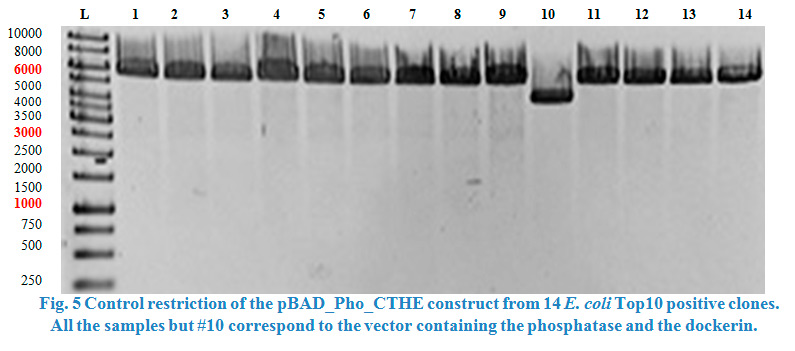
| + | <p> | ||
| + | Restriction control was done with the enzymes SacI and PvuII from Thermo fisher scientific.<strong> </strong>Restricted plasmids were checked on a 0.8 % | ||
| + | agarose gel. | ||
| + | </p> | ||
| + | </html> | ||
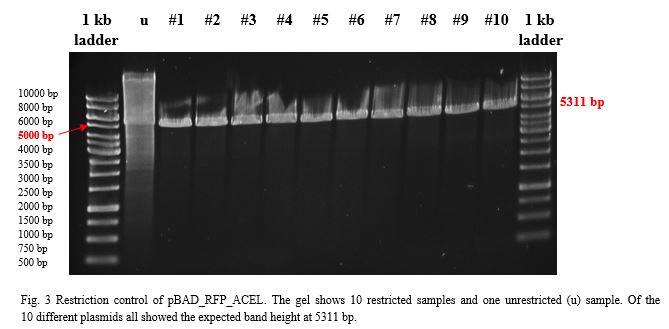
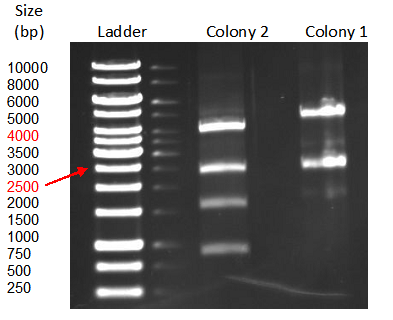
| + | [[File:TeamGoettingen_Restriction_pBAD_CellulaseCCEL.jpg|300px|center|thumb|Figure 3: Restriction control of the pBAD_CellulaseCCEL construct from TOP10 E. coli positive clones by with SacI and PvuII. Colony 1 has the desired insert.]] | ||
| + | <html> | ||
| + | <p> | ||
| + | The correct insert was confirmed through sequencing. | ||
| + | </p> | ||
| + | </div> | ||
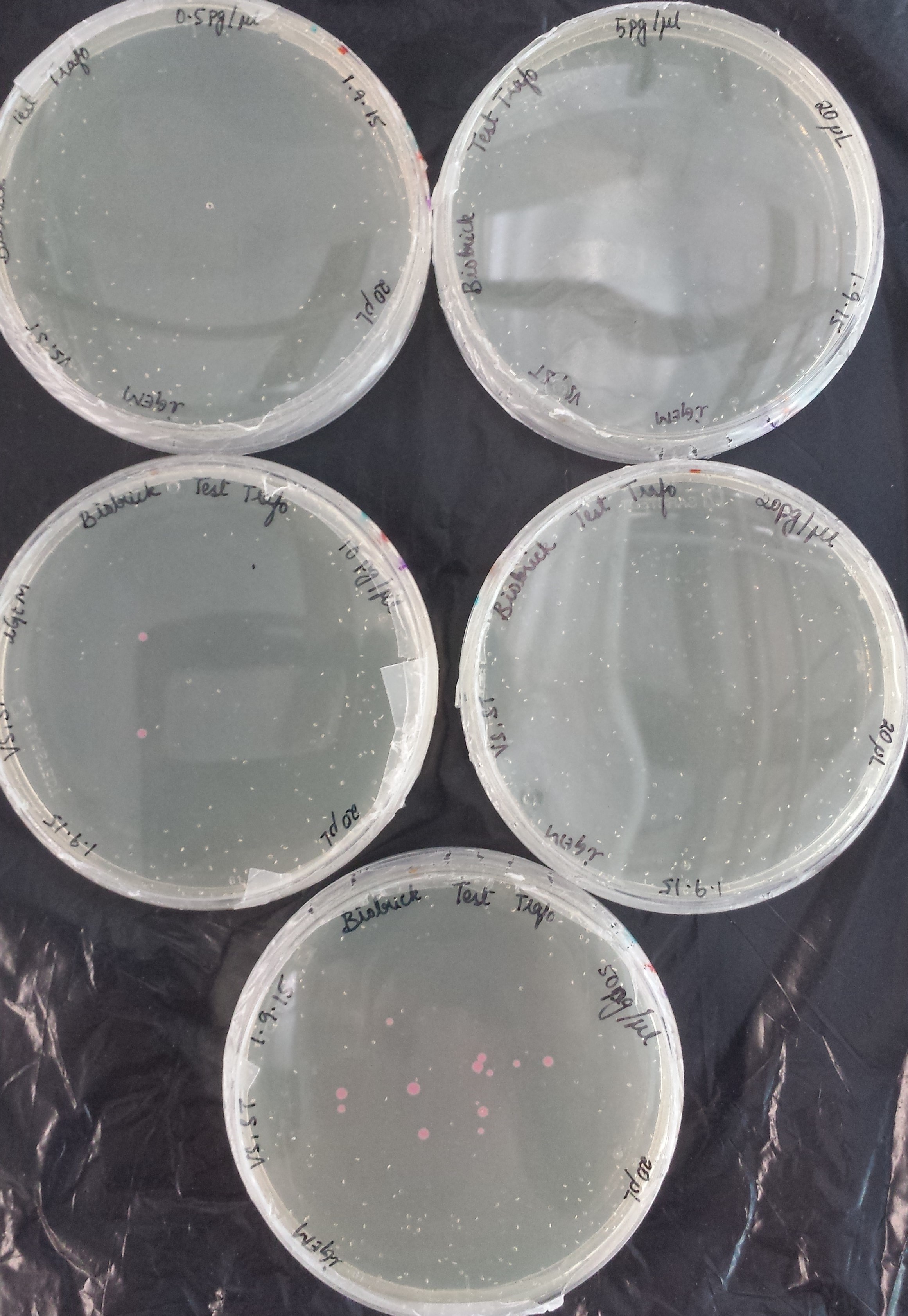
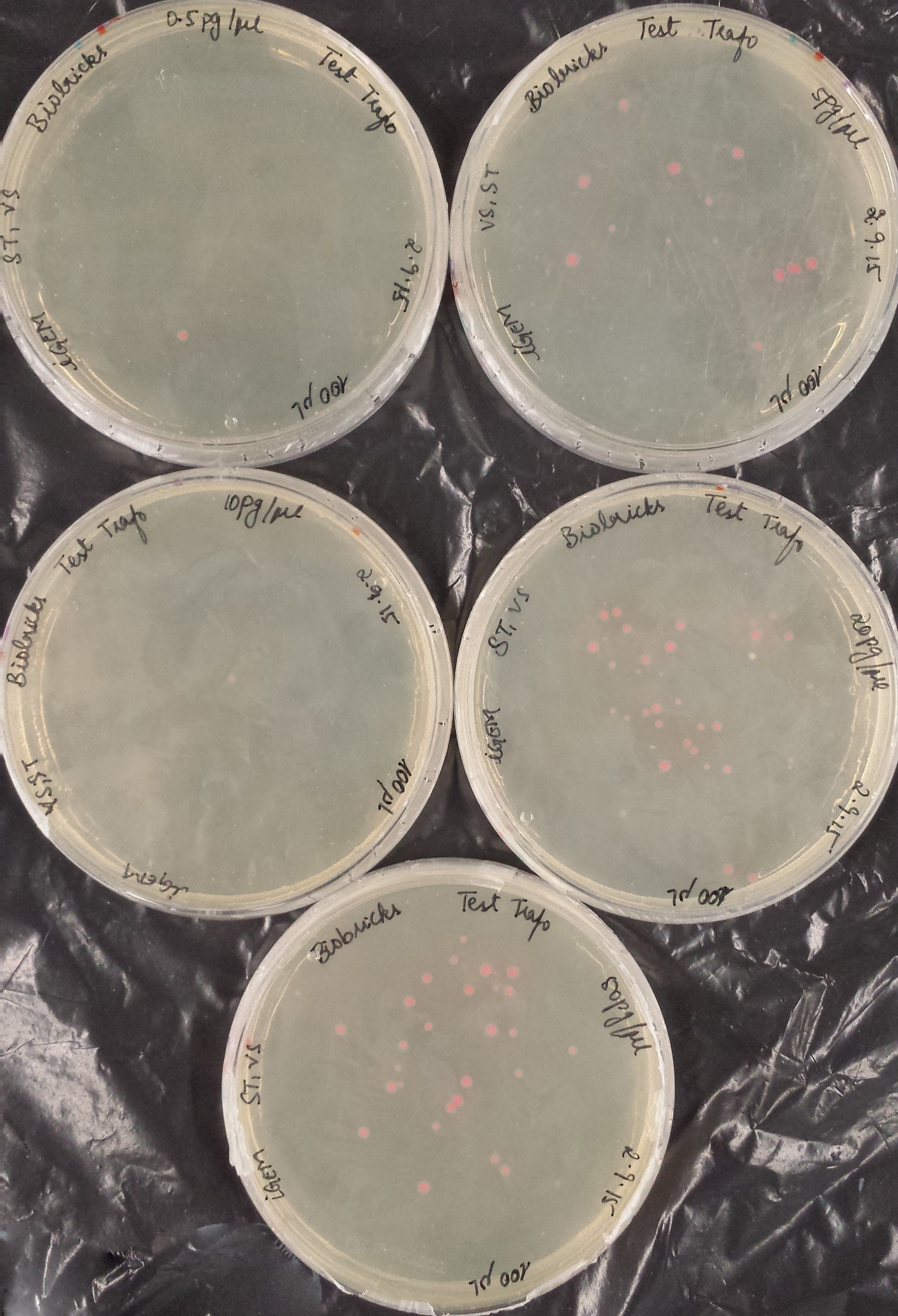
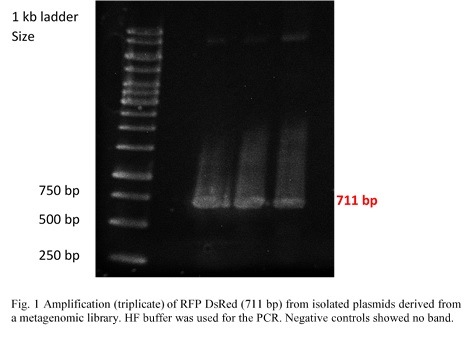
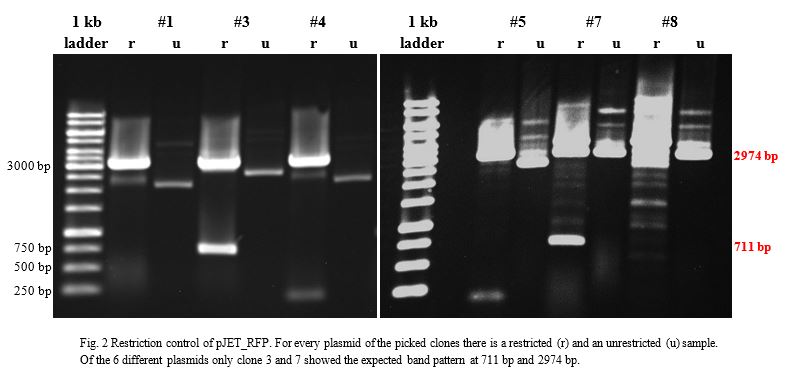
<a href="" onClick=" $('#menu4').slideToggle(300, function callback() { }); return false;"><h1>BioBricks</h1></a> | <a href="" onClick=" $('#menu4').slideToggle(300, function callback() { }); return false;"><h1>BioBricks</h1></a> | ||
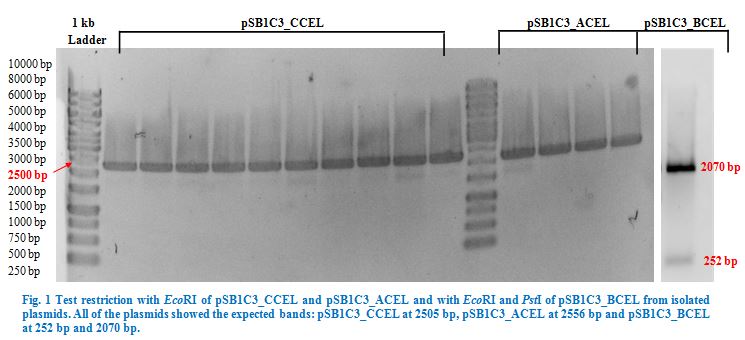
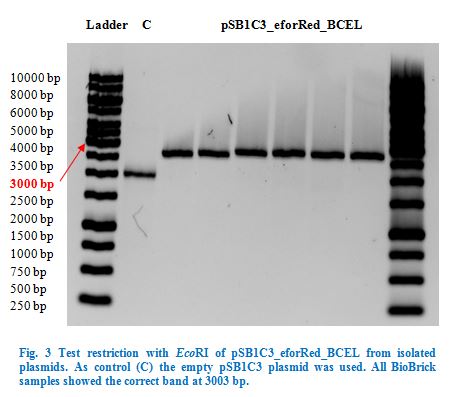
| Line 392: | Line 458: | ||
Once restriction controls showed the correct bands, pSB1C3_CCEL, pSB1C3_ACEL and pSB1C3_BCEL were sent for sequencing. | Once restriction controls showed the correct bands, pSB1C3_CCEL, pSB1C3_ACEL and pSB1C3_BCEL were sent for sequencing. | ||
</p> | </p> | ||
| + | <p> </p> | ||
<p> | <p> | ||
| + | |||
<strong>Colour BioBricks</strong> | <strong>Colour BioBricks</strong> | ||
</p> | </p> | ||
<p> | <p> | ||
| − | We also wanted to improve already existing BioBricks by fusing our dockerins to the colours eforRed (BBa_K592012) and amilCP (BBa_K592009) that were | + | We also wanted to improve already existing BioBricks by fusing our dockerins to the colours eforRed (<a href="http://parts.igem.org/Part:BBa_K592012"style="color: #000066">BBa_K592012</a>) and amilCP (<a href="http://parts.igem.org/Part:BBa_K592009"style="color: #000066">BBa_K592009</a>) that were |
submitted from the University of Uppsala (Sweden) in 2011. | submitted from the University of Uppsala (Sweden) in 2011. | ||
</p> | </p> | ||
| Line 455: | Line 523: | ||
</p> | </p> | ||
<p> | <p> | ||
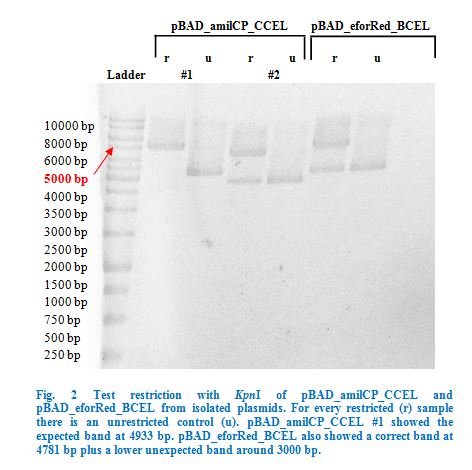
| − | Furthermore we collaborated with the current iGEM team of Aachen and tried to fuse three of their enzymes to our constructs | + | Furthermore we <a href="https://2015.igem.org/Team:Goettingen/Collaborations"style="color: #000066">collaborated</a> with the current iGEM team of Aachen and tried to fuse three of their enzymes to our constructs but could not finish our work here. |
| − | + | ||
</p> | </p> | ||
| + | |||
<p> | <p> | ||
<strong>ACHIEVENTS<a name="_GoBack"></a></strong> | <strong>ACHIEVENTS<a name="_GoBack"></a></strong> | ||
| Line 465: | Line 533: | ||
</p> | </p> | ||
<p> | <p> | ||
| − | pSB1C3_BCEL ( | + | pSB1C3_BCEL (<a href="http://parts.igem.org/Part:BBa_K1865000"style="color: #000066">BBa_K1865000</a>) |
</p> | </p> | ||
<p> | <p> | ||
| − | pSB1C3_eforRed_BCEL ( | + | pSB1C3_eforRed_BCEL (<a href="http://parts.igem.org/Part:BBa_K1865001"style="color: #000066">BBa_K1865001</a>) |
</p> | </p> | ||
| + | </div> | ||
| + | |||
| + | <a href="" onClick=" $('#menu6').slideToggle(300, function callback() { }); return false;"><h1>Scaffoldin Purification</h1></a> | ||
| + | <div id="menu6"> | ||
| + | <p> | ||
| + | <strong>Scaffoldin purification</strong> | ||
| + | </p> | ||
| + | <p> | ||
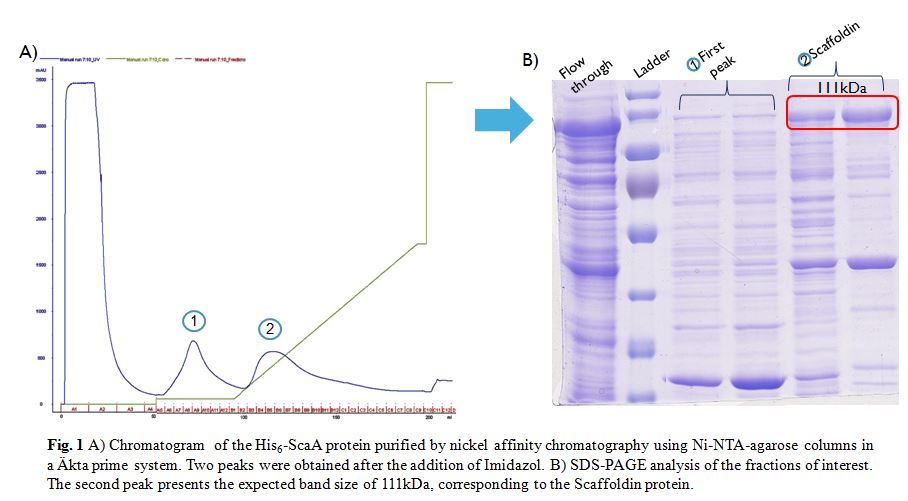
| + | After induction according to the methods collection, the targeted ScaA scaffoldin was purified by nickel affinity chromatography using Ni-NTA-agarose columns in a Äkta prime system (Fig. 1A). Following the addition of Imidazol two peaks were obtained. The fractions of interest were visualized in a SDS-PAGE gel (Fig. 1B). The gel showed that only the second fraction presents the expected size of 111kDa. However some other bands appeared along this fraction, therefore in order to obtain even more pure protein a Size Exclusion Chromatography (SEC) was done. | ||
| + | </p> | ||
| + | </html> | ||
| + | [[File:Histag_purification_iGEM_Goettingen2015.jpeg|600px|thumb|center|]] | ||
| + | <html> | ||
| + | <p> | ||
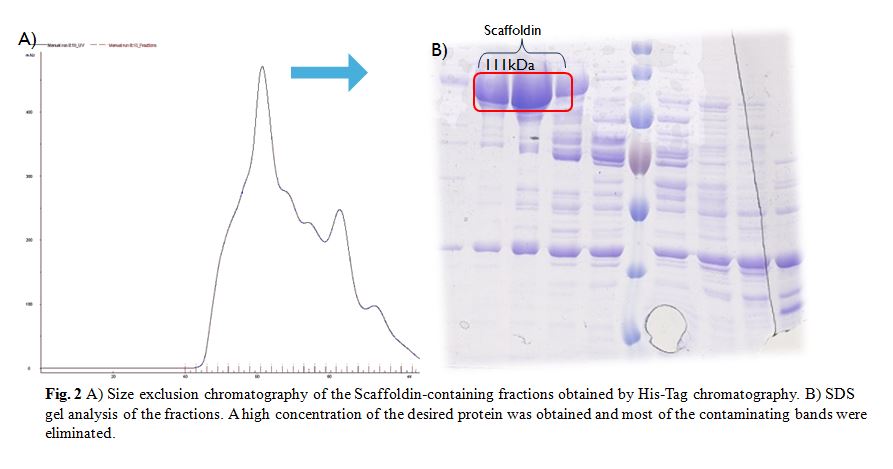
| + | The SEC was done with a HiLoad Superdex 200 16/600 column (GE Healthcare) (Fig. 2A). SDS gel shows that a high concentration of the desired protein was | ||
| + | obtained and most of the previous contaminating bands were eliminated. Nevertheless there was an unknown band of low weight present in every fraction, | ||
| + | apparently it could be a second protein from E. coli with some affinity to the Scaffoldin than is then is co-purified with it (Fig. 2B). | ||
| + | </p> | ||
| + | </html> | ||
| + | [[File:SEC_iGEM_Gottingen2015.jpeg|600px|thumb|center|]] | ||
| + | <html> | ||
| + | <p> | ||
| + | Future plans include to incubate the Scafoldin with the purified enzyme-dockerin construct in order to prove that the Flexosome can assembly by itself. | ||
| + | </p> | ||
| + | </div> | ||
| + | <a href="" onClick=" $('#menu7').slideToggle(300, function callback() { }); return false;"><h1>Copurification</h1></a> | ||
| + | <div id="menu7"> | ||
| + | <p> | ||
| + | Copurification of Scaffoldin and Enzyme-Dockerin | ||
| + | </p> | ||
| + | <p> | ||
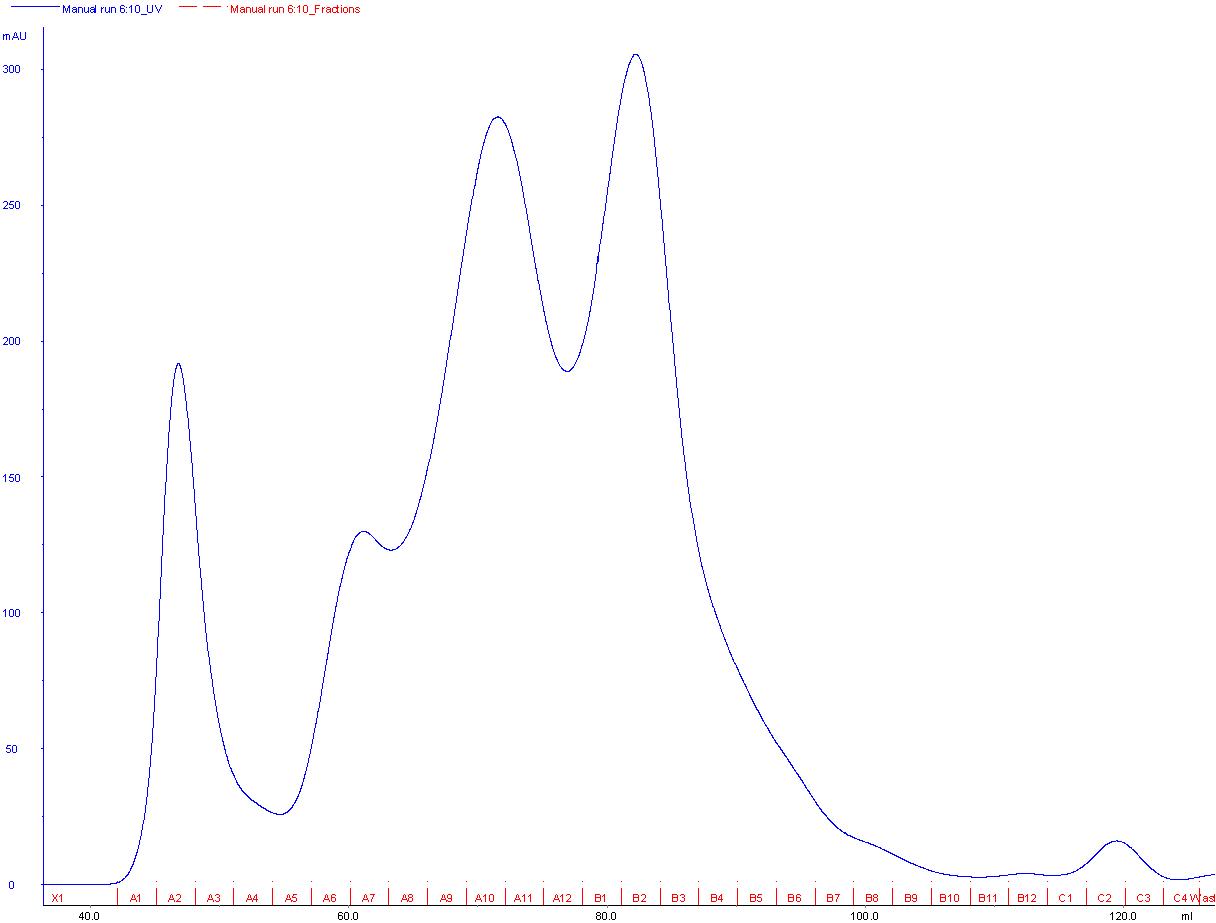
| + | The main peaks after gelfiltration from the scaffoldin CTHE (see results scaffoldin) and CellulaseCCEL (Fig.1) were pooled and concentrated. | ||
| + | </p> | ||
| + | </html> | ||
| + | [[File:TeamGoettingen_GeFi_CellulaseCCEL.jpg|650px|centre|thumb|B1-B3 were pooled for copurification.]] | ||
| + | <html> | ||
| + | <p> | ||
| + | They were then mixed and incubated together at 37°C for 15 min with agitation. | ||
| + | </p> | ||
| + | <p> | ||
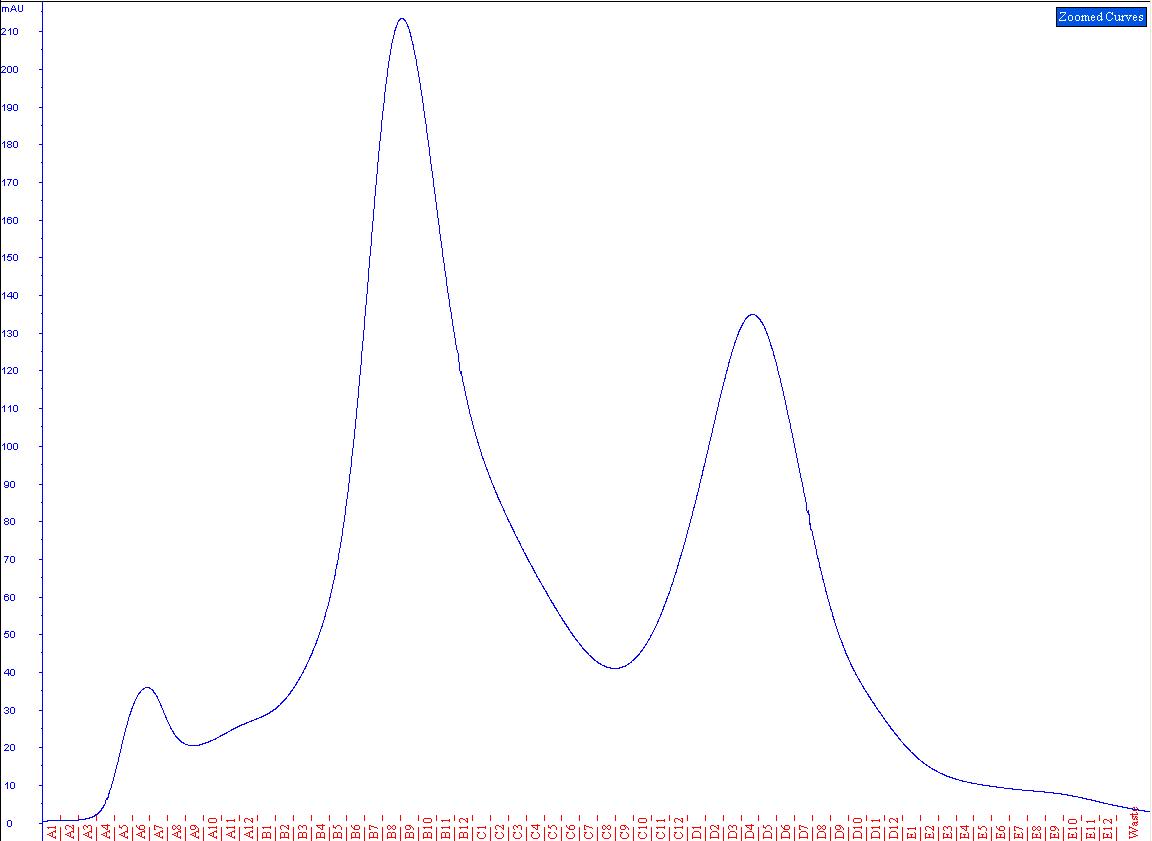
| + | The solution was then used in another gelfiltration (Fig.2). The resulting peaks were run on a SDS-gel (Fig.3) and checked for cellulase activity. | ||
| + | </p> | ||
| + | </html> | ||
| + | [[File:TeamGoettingen_Copurification_CTHE_CellulaseCCEL.jpg|650px|centre|thumb|Fig.2) The peak at A5-A7 may be the bound scaffoldin and dockerin, whilst the unbound proteins are expected between B3 and C9.]] | ||
| + | |||
| + | <html> | ||
| + | <p> | ||
| + | The first peak from the copurification showed a distinct cellulase activity. As the peak ran considerably higher than expected than either the scaffoldin | ||
| + | or the CellulaseCCEL in the gelfitration, it can be expected that this peak contains a successfully formed complex between CellulaseCCEL and the CTHE | ||
| + | scaffoldin. | ||
| + | </p> | ||
| + | </div> | ||
| + | <a href="" onClick=" $('#menu8').slideToggle(300, function callback() { }); return false;"><h1>Giant Jamboree: Poster and Presentation</h1></a> | ||
| + | <div id="menu8"> | ||
| + | <p> | ||
| + | Did you missed our presentation? Do you want to see our poster again? | ||
| + | No problem, here you can find the files: | ||
| + | </p> | ||
| + | <p> | ||
| + | </html> | ||
| + | [[Media:IGEM-presentation_final_team_Goettingen.ppt]] | ||
| + | <html> | ||
| + | </p> | ||
| + | <p> | ||
| + | </html> | ||
| + | [[Media:Poster_teamGoettingen_2015.pdf]] | ||
| + | <html> | ||
| + | </p> | ||
</div> | </div> | ||
</html> | </html> | ||
Latest revision as of 22:03, 20 October 2015