Team:Austin UTexas/Project/Problem
Problem: Genetic Instability
Background
One of the major issues facing genetic engineering is the longevity of genetic devices once inserted into organisms. An organism can be modified, but that does not ensure that the modification will last. Subsequent generations of modified organisms often lose or “break” the genetic device through mutations. Breaking a plasmid or an inserted device often decreases the metabolic load of that organism, giving it a competitive advantage by enabling it to allocate more resources toward replication. This allows them to dominate a population over time.
Previous research has identified several factors that contribute to the breaking of genetic devices (1). One major factor is the relative fitness of organisms without the device over those with it. Strongly expressed devices will result in a broken population faster than weakly expressed devices, since devices of the former type place a greater metabolic load on the organism. Similarly, genes that code for 'costly' proteins that may interfere with other cellular functions in ways that are toxic are more likely to have a broken device sweep through the populaiton.
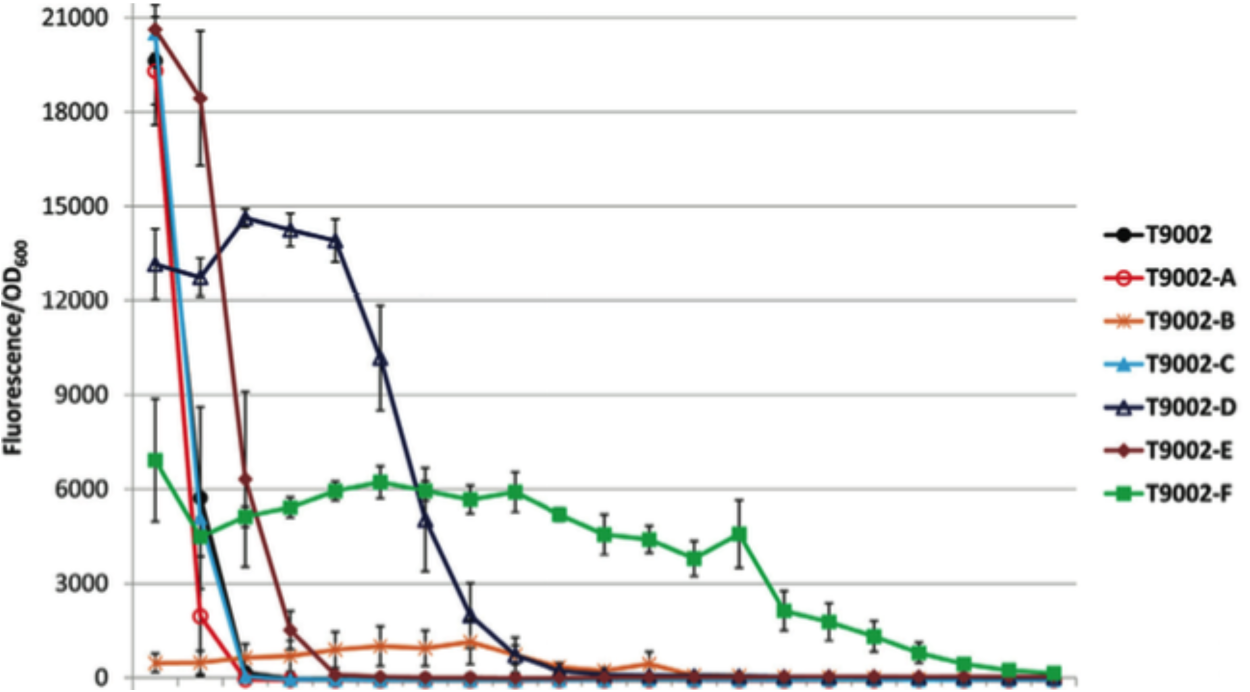
Another major factor that contributes to device stability is the rate of mutations leading to device failure. If a device includes DNA sequences that act as mutational hotspots, it may be very unstable. The [http://barricklab.org/django/efm Evolutionary Failure Mode calculator] (developed by the Barrick lab) identifies DNA sequences that are prone to breaking, such as simple sequence repeats and long stretches of a single repeated nucleotide. The EFM calculator gives sequences a relative instability prediction (RIP) score, and higher RIP scores indicate sequences that are more prone to breaking.
Project and Motivation
Our projects focused on monitoring device stability and determining what kinds of sequences and sequence features contribute to device instability. We wish to identify and better characterize sequences that contribute to the relative instability of genetic devices so they can be used to predict device longevity, as well as be taken into account when creating and modifying such devices. In the spring of 2015, UT iGEM team members, assisted by the UT Austin Freshman Research Initiative stream "Hijacking Cell Factories for Synthetic Biology," utilized the EFM calculator to identify sequences within fluorescent protein coding genes that were prone to mutation. We used this information to determine which coding sequences were most and least evolutionary stable. Then, over the summer we chose one of the unstable coding sequences and tested its stability in different strains of E. coli. Further, we redesigned the pDCAF3 plasmid ([http://parts.igem.org/wiki/index.php?title=Part:BBa_K734000 BBa_K734000]) made by the 2012 Austin UTexas iGEM team in order to replace potentially unstable sequences while retaining the original functionality. In addition, we designed a set of plasmids to expand on the use of the pDCAF3 plasmid for measuring caffeine levels in beverages.
A Computational Approach
In the course of our experiments with genetic device stability, we also investigated ways to use the power of computation to study our devices. Results gathered using these methods provided similar data, yet offered efficiency improvements and stronger part verification over human analysis. Such advances are key for scaling up the project to higher throughput technologies such as flow cytometry and Illumina sequencing. Higher throughput will allow us to generate better statistics for determining how quickly a device breaks.
Continue to Part 1: OBSERVING FLUORESCENT GENE STABILITY
References
- Sleight et al.: Designing and engineering evolutionary robust genetic circuits. Journal of Biological Engineering 2010 4:12.
- “EFMCalculator.” tool from the Barrick Lab: http://barricklab.org/django/efm/
- Jack et al.: Predicting the genetic stability of engineered DNA sequences with the EFM calculator. Print. Sept. 2015