Team:Brasil-USP/Parts/Teamparts
Team Parts

Favorite Parts
RoxA - Rubber Oxygenase A
Part BBa_K1819003 corresponds to the coding sequence of rubber oxygenase A (Rox A) from Xanthomonas sp. strain 35Y (Gene bank accession code: AGT20506.1). This 73 kDa protein is an extracellular enzyme that catalyzes oxidative C-C cleavage of poly(cis-1,4-isoprene) to a major product, 12-oxo-4,8-dimethyltrideca-4,8-diene-1-al (ODTD; C15 tri-isoprenoid), with specific activity measured as 0.3 mol min-1 per mg.

Found not only in Xanthomonas sp. but also in other Gram-negative clearing zone formers, RoxA is structurally related to cytochrome c peroxidases and its function is associated to two covalently bound heme groups. Although recently characterized by Birke et al1, the exact enzymatic mechanism of rubber degradation by RoxA has not yet been elucidated.

Because Xanthomonas sp. RoxA is exported to the extracellular environment, its open reading frame contains a sequence that codes to a signal peptide flanking its 5’-terminus. We detected this region (amino acid residues from 1 to 20) using signal peptide predictor server4 (www.compgen.org/tools/PRED-TAT) and eliminated it to generate part BBa_K1819003 with the coding sequence of RoxA flanked by SacI and NdeI restriction sites. Such non-usual restriction sites may serve for cloning purposes that lead to expression of recombinant protein with no additional amino acid residue(s) between its coding sequence and the fused N-terminal tag. Part BBa_K1819003 was cloned in pSB1C3 plasmid and submitted to Registry of Standard Biological Parts.

Lcp - Latex Clearing Protein
Part BBa_K1819001 corresponds to the coding sequence of latex clearing protein (Lcp) from actinomycete Streptomyces sp. strain K30 (GenBank accession code: AAR25849 colocar link ). Other Gram-positive clearing zone bacteria also present this 42 kDa enzyme, which catalyzes rubber cleavage at the cis double bonds to multiple products ranging from C20 tetra-isoprenoid to at least C35 hepta-isoprenoid with measured specific activity of 1.3 mol min-1 per mg. Lcp contains an oxidized heme-Fe3+ not bound to dioxygen as in RoxA 2. Neither its structure nor its catalysis mechanism were determined until now.

Again, because Lcp is secreted by Streptomyces sp., it contains a 5’-signal peptide that was identified using signal peptide predictor server4 (www.compgen.org/tools/PRED-TAT). We removed this region from Lcp to generate part BBa_K1819001 flanked by SacI and NdeI restrictions sites. We also submitted it to the Registry.
Linker+GFP
To characterize expression and secretion of mature proteins, we designed a new part that combines GFP coding sequence with an N-terminal linker, which is 5’-flanked by NdeI restriction site. Such part may serve to other IGEM teams that would like to express a chimera C-terminally attached to GFP, which exhibits bright green fluorescence when exposed to light in the blue to ultraviolet range.
This part can be especially helpful in characterization of gene expression and parameter fitting, as we describe in our modeling section. It is, however, mandatory the addition of a Ribosomal Binding Site (RBS) before the protein linked in the linker+gfp genes. As shown in figure 6, missing the RBS simply produce background fluorescence as the protein linked to GFP will not be translated.
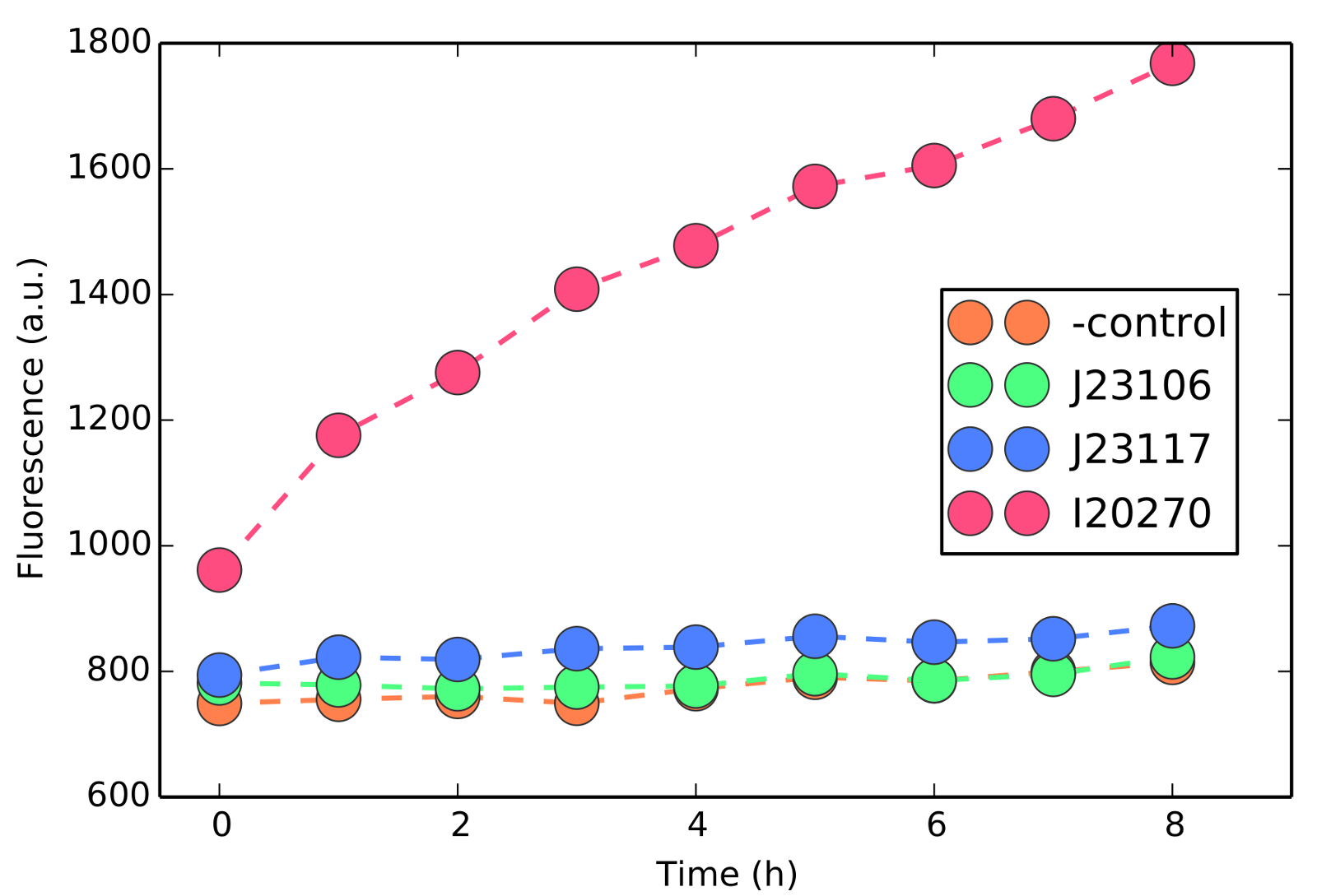
Figure 6- Usage of BBa_K1819000 without adding a Ribosomal Binding Site (RBS) before the linker+gfp genes. Gfp proteins will simply not be translated and no green signal is observed other than the background fluorescence (negative control).
TAT signal - Twin-arginine translocation signal
Part BBa_K1819002 corresponds to a Twin-arginine translocation signal (TAT signal), which is 3’-flanked by SacI restriction site. We believe that other IGEM teams may want to use TAT signal from E. coli in their own projects. Therefore, after cloning into pSB1C3 plasmid and sequencing, we will submit TAT signal coding region of hydrogenase-1 small chain precursor from E. coli as a new part in biobrick format (part BBa_K1819002). SacI restriction site will be add at the 3’ flanking region of TAT signal to link it at the first amino acid residue of the protein to be exported.

References
1. Birke J, Jendrossek D. Rubber oxygenase and latex clearing protein cleave rubber to different products and use different cleavage mechanisms. Applied and Environmental Microbiology. 2014;80(16):5012-5020.
2. Birke J, Röther W and Jendrossek D. Latex Clearing Protein (Lcp) of Streptomyces sp. Strain K30 Is a b-Type Cytochrome and Differs from Rubber Oxygenase A (RoxA) in Its Biophysical Properties. Applied and Environmental Microbiology. 2015; 81(11):3793-3799.
3. Hiessl S, Böse D, Oetermann S, Eggers J, Pietruszka J, Steinbüchel A. Latex Clearing Protein—an Oxygenase Cleaving Poly(cis-1,4-Isoprene) Rubber at thecis Double Bonds. Kivisaar M, ed. Applied and Environmental Microbiology. 2014;80(17):5231-5240.
4. Bagos PG, Nikolaou EP, Liakopoulos TD, Tsirigos KD. Combined prediction of Tat and Sec signal peptides with Hidden Markov Models. Bioinformatics. 2010; 6(22):2811-2817.
5. Seidel J, Schmitt G, Hoffmann M, Jendrossek D, Einsle O. Structure of the processive rubber oxygenase RoxA from Xanthomonas sp. Proceedings of the National Academy of Sciences USA. 2013;110(34):13833-13838.
6. Yikmis, M. et al. Secretion and transcriptional regulation of the latex-clearing protein, lcp, by the rubber-degrading bacterium Streptomyces sp strain K30. Appl. Environ. Microbiol. 74, 5373-5382, doi:10.1128/aem.01001-08 (2008).


