Team:UMBC-Maryland/Description
Project Description
Copper is a major pollutant in a variety of fresh water ecosystems. When copper is oxidized from Cu+ to Cu2+, it often produces a free radical known as a reactive oxygen species (ROS) which is capable of severely damaging any biological molecules it comes into contact with. This damage at the molecular level has been linked to diseases such as Parkinson's, Alzheimer's, and Prion diseases. E. coli have been engineered to have increased copper uptake; however, due to the toxicity of copper, the E. coli quickly saturate and are unable to uptake any more copper. Our goal is to increase the efficiency of copper uptake in E. coli for the purpose of bioremediation in fresh water ecosystems. We plan on doing this by increasing our engineered cells' resistance to copper toxicity as well as increasing their ability to uptake copper.
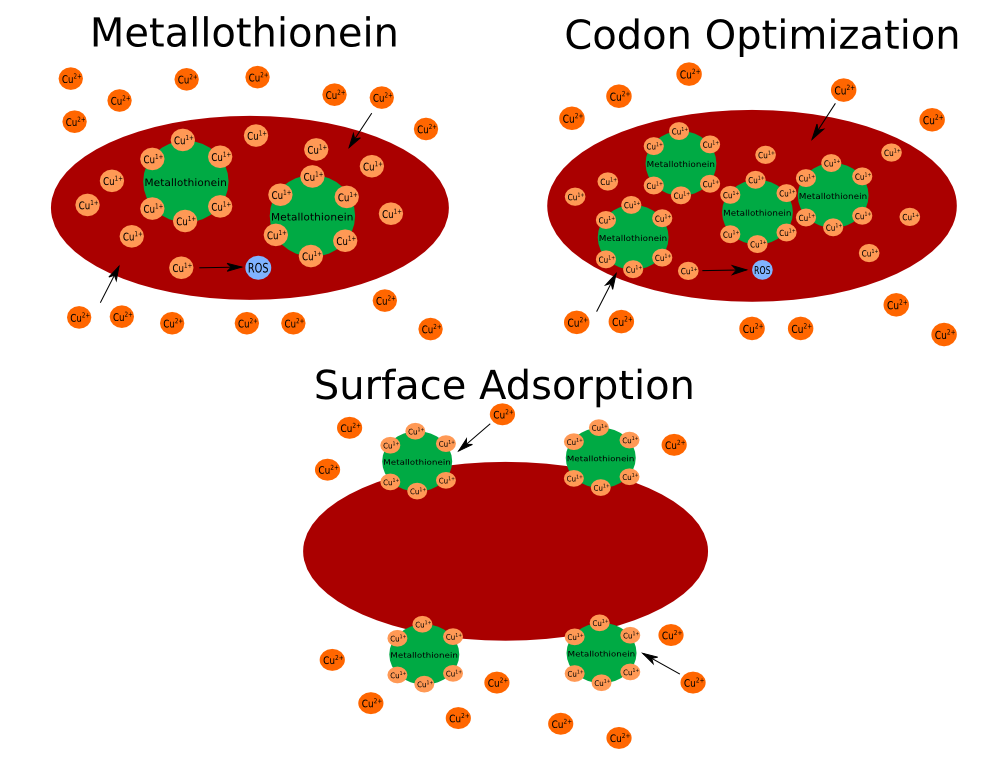
In order to make engineered cells more resistant to copper toxicity, we will be genetically engineering E. coli with enhanced copper tolerance by giving it a gene for yeast metallothionein. This protein will bind 11 copper atoms, making them less reactive and less toxic. This protein is also an antioxidant that can detoxify hydroxyl radicals with its cysteine groups.
We plan on creating three separate cell lines that contain a metallothionein gene in different forms. One is the yeast metallothionein CUP1 gene directly transformed into E. coli. A second is the CUP1 gene codon optimized for E. coli. The third is metallothionein that will adhere to the outer membrane of E. coli. This will be done by fusing the CUP1 gene sequence into the lamB transport channel. This will cause metallothionein to adhere to the outer surface of E. coli by being bound to one of E. coli’s outer membrane transporters. This should ideally allow engineered cells to be able to sequester copper without getting as much inside, making copper less toxic to them.
Future efforts to bring this project to its complete application in polluted waters will include several additional genes. Superoxide dismutase and catalase are integral antioxidant proteins that are key to radical scavenging. Without them, ROS will continue to cause cell damage as quickly as heavy metals like copper will produce ROS. Superoxide dismutase converts superoxide ROS molecules into hydrogren peroxide. Catalse converts hydrogen peroxide into water. In addition to these antioxidants, a buoyancy gene under the control of a copper sensitive promoter can be utilized to gather and collect engineered cells. The use of these combined tools will aid in the harvesting of heavy metals such as copper from polluted waters.

