Team:UMBC-Maryland/Notebook
Notebook
Week 1
June 1st
First meeting of the project over summer where we ran our PCR products on a 1% agarose gel for practice.
June 4th
iGEM team members attended a meeting with a professor and was given an overview to the procedure of 3A Assembly. We also discussed team structure and the CUP1 gene construct that is needed to be synthesized as a G-block. G-blocks are then submitted to IDT for synthesis.
Week 2
June 8th
Digest products with a combination of restriction enzymes (XbaI, HindIII, EcoRI)were ran on a 2% agarose gel. Both gel results came up inconclusive.
Week 3
June 15th
Repeated digestion to troubleshoot any possible errors that may have occurred during the protocol.
Week 4
June 22nd
Repeated analysis of the digestion products on the gel. Gel results showed no bands
June 25th
Mini-prep of the metallothionein plasmid.
Week 5
June 29th
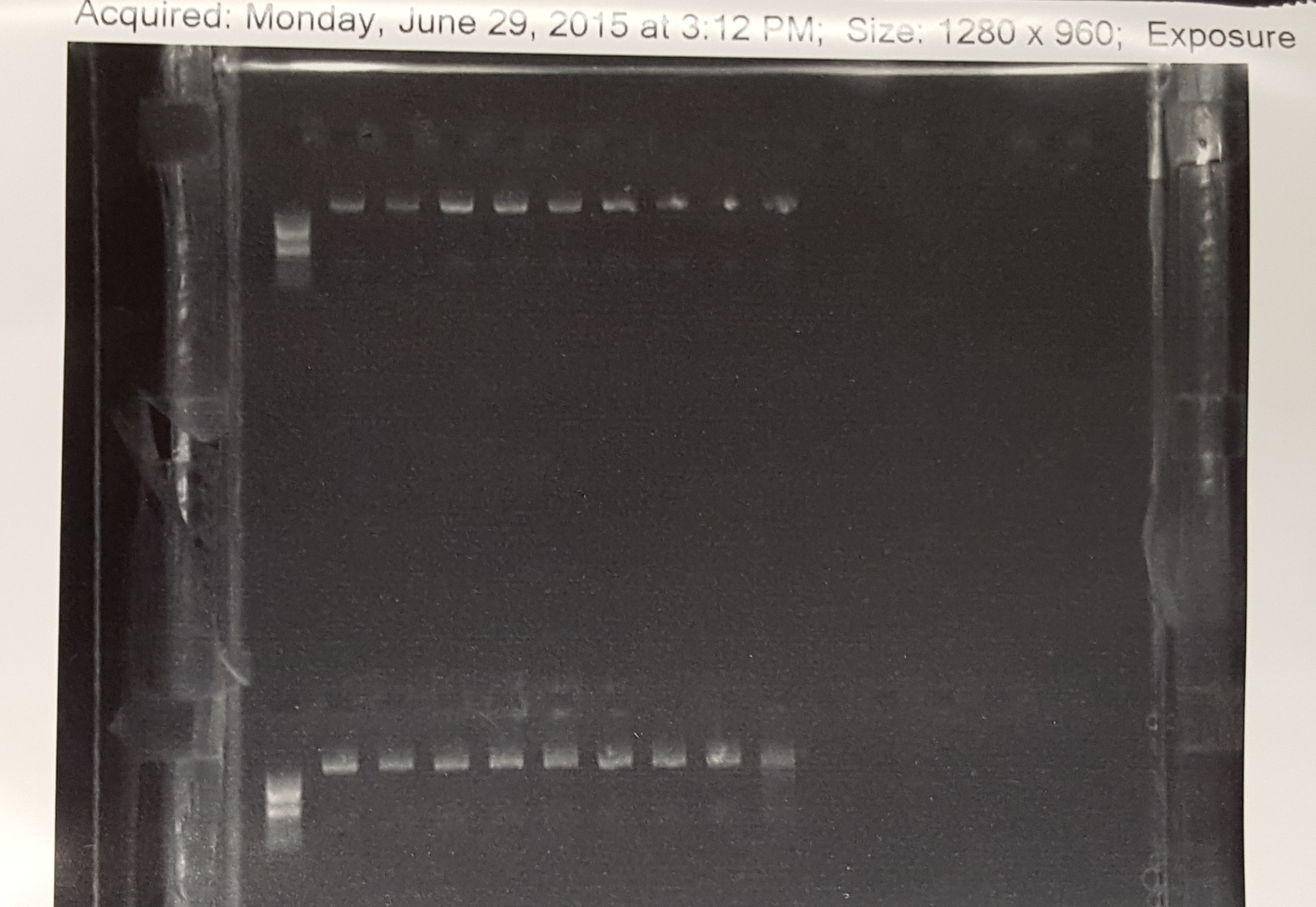
Digestion of the metallothionein plasmid with restriction enzymes (EcoRI & PstI). After further investigation, the Bulls-Eye solution was the cause of bands not appearing and we switched to Ethidium Bromide. The digestion products were then run on a gel stained with ethidium bromide. The gel depicted both digested and undigested bands that can be used to identify whether our restriction enzymes are working or not.
Week 6
July 7th
Today marks the first day we began production of our first cell line, Retriever 1(Ret1).The CUP1 transformed cells and control cells were plated and an overnight was done.
July 8th
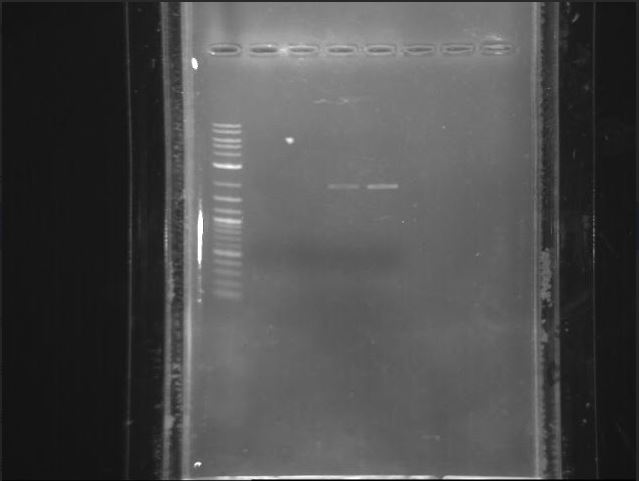
A gel was run on the transformed cells and control cells using Bullseye (left) and EtBr (right) to stain the DNA. We concluded that we would be using EtBr thereafter.
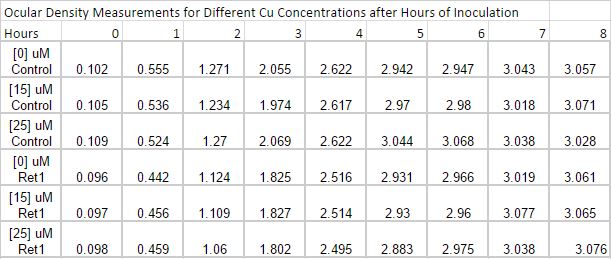
July 9th
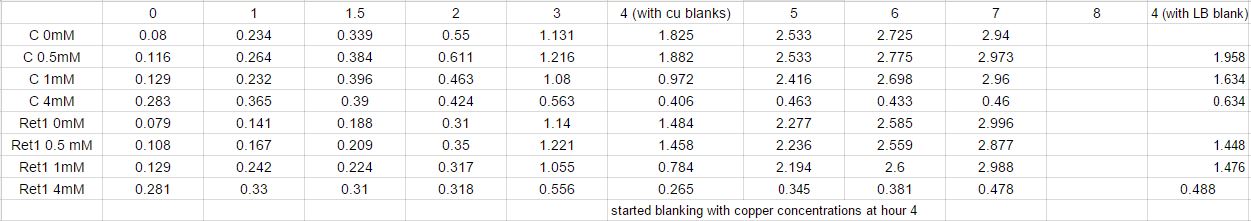
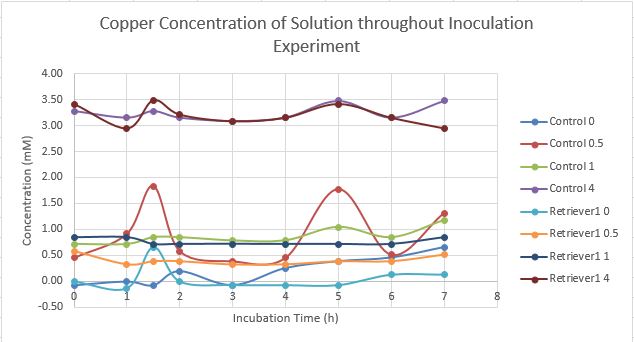
Inoculation experiment. Six flasks of 25 mL LB broth were prepared, 3 of which will be the control and the other will be for the transformed. From Copper Sulfate, a stock solution of 0.5M Copper Solution was created and then a 0.1M Solution was created to be used. Enough volume of the 0.1M Copper Solution was added to the 6 six flasks so each of the control and transformed group will have flasks containing a concentration of [0uM], [15uM] and [25uM]. To the transformed group 9.375uL of Chloramphenicol was added. At every hour for eight hours, starting at the 0th hour, an optical density reading will be taken for each flask. Drawing out 1mL of the solution from each flask into eppendorfs, followed by measuring the optical density. Since the Laemmli Buffer was nowhere to be found,the solution was spun down to be put in the freezer for the SDS gel in the future. Another 1 mL from each flask is saved for copper measurement. Data of time points is displayed below.
Week 7
July 15th
Meeting with our advisor regarding current updates on the project and the availability of the flame atomic absorption spectrometer to be used for the copper measurement. An overnight for the Ret1 transformed cells was done today to repeat the experiment at higher concentrations of copper.
July 16th
Another inoculation experiment was scheduled for today, however the tube of the overnight cells broke so instead a repeat of the overnight was done.
July 17th
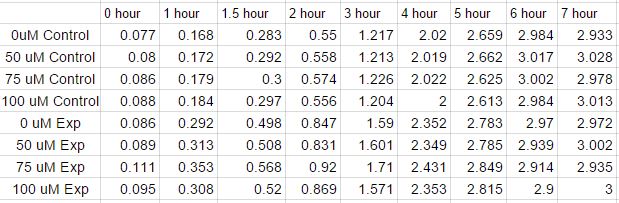
The second inoculation experiment. A copper concentration of [0uM], [50uM], [75uM] and [100uM] was experimented with. Data was collected in the same manner except with a 30 minute time point after the one hour mark. After the optical density was read for the solution, the solutions were centrifuged and the supernatant in the eppendorfs were removed to be used for copper measurement. The pellet in the eppendorfs were then resuspended with 200uL of master mix of Laemmli Buffer and were placed in the freezer to be used to run an SDS-gel in the future.
Week 8
July 21st
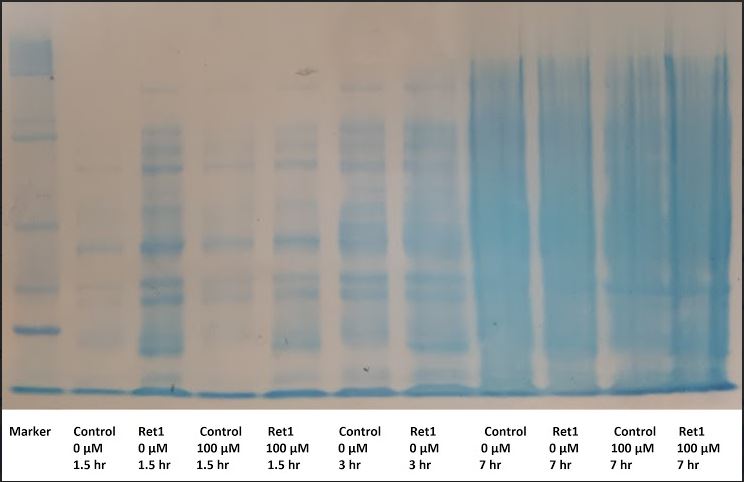
From the second inoculation experiment, the eppendorfs were thawed on ice in preparation for the 10% SDS-gel. 7 uL of the Sigma Aldrich Wide Range Ladder (6-205 kDA) was added to the first lane while the other 10 lanes each contained 10 uL of samples from the experiment. Samples at timepoints 1.5, 3, 7, were selected at different concentrations. The gel was ran at 125V for 40 minutes and then stained with Blue Staining Solution. The gel was decanted for an hour with repeated rinses every 15 minutes. gel results are discussed on a later date.
Week 9
July 27th
Discussion of the gel results and future plans on how copper measurement will be done. We planned to use flame atomic absorption spectroscopy, however our copper concentrations were too small. The second method uses a plate reader to measure the florescence of the sample.
Week 10
July 31st
Made BCSsolution for detection of copper solution composition of single sample 4ul of copper sample 10ul of 10 uM reducing agent (sodium citrate) 10 ul of 4uM BCS 76 ul of PBS
August 3rd
The flame atomic absorption spectroscopy was not available for use since the instructor was on vacation so we used the plate reader. The plate reader could not measure the florescence because there were no appropriate filters in the lab to emit higher than 700nm. Instead, we looked toward other options for copper measurement.
August 6th
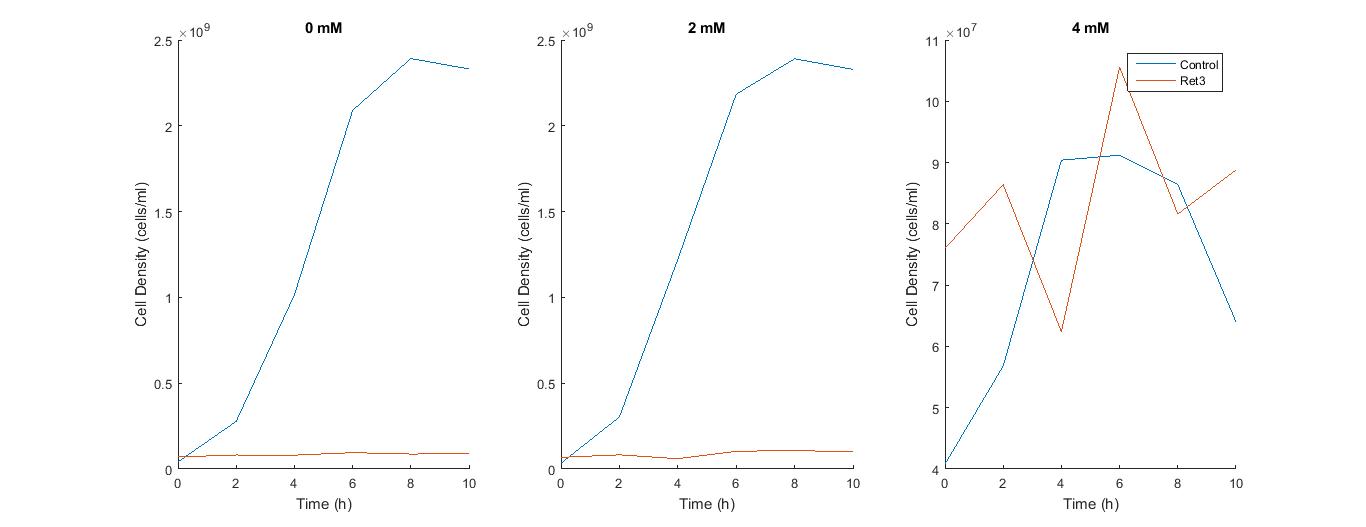
The third growth experiment was scheduled today. We worked with concentrations of 0 mM, 0.5 mM, 1 mM, and 4mM. We noticed that we had to create different blanks due to the different copper concentrations that we did not do for the previous experiments. Samples were then stored for SDS-gels again. The second iGEM part, CUP1 codon optimized came in the day prior. Hi-Fi assembly was performed followed by transformation. The control was spread on plates with ampicilin resistance and the transformed group was spread on plates with chloramphenicol resistance. Each group contained plates consisting of 25 uM, 50 uM, and 100 uM. The plates were incubated and grown overnight.
Week 11
August 11th
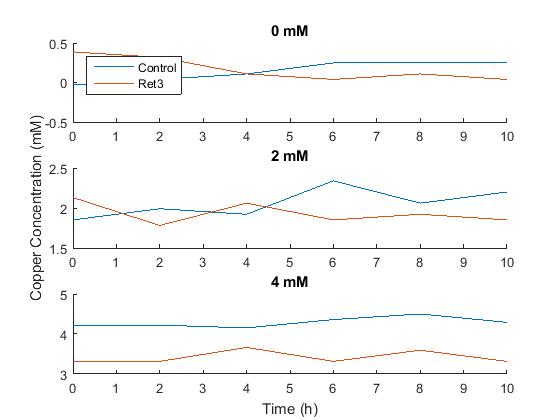
The control plate had only a few colonies growing on it, even after a second attempt while the transformed had multiple colonies. We currently do not have an explanation for why there are less colonies on the control plate, however we decided to do a miniprep of the CUP1 codon optimized gene in order to do the restriction digest and gel the next experiment day. Copper measurements were taken through the use of a different plate reader. The absorbance of copper was taken at all the time points and a graph was created depicting the data.
August 13th
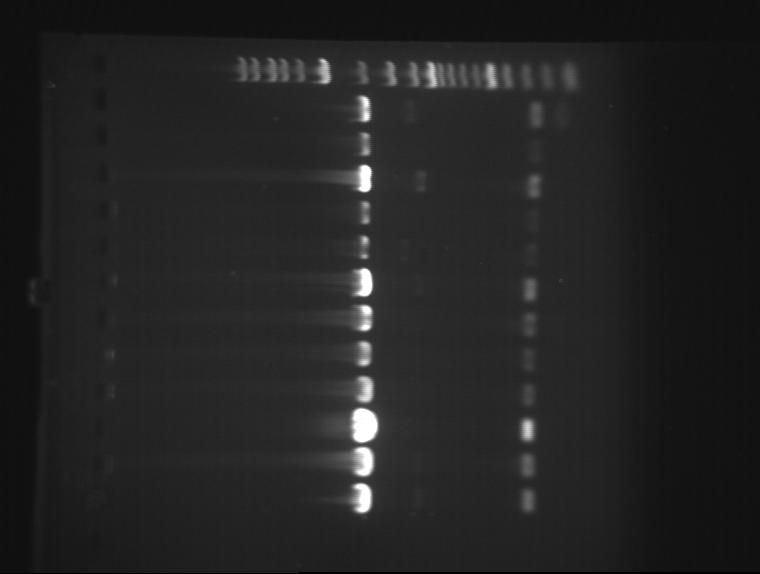
The DNA was digested with PstI and EcoRI and then ran on a 1% agarose gel.
Week 12
August 17th
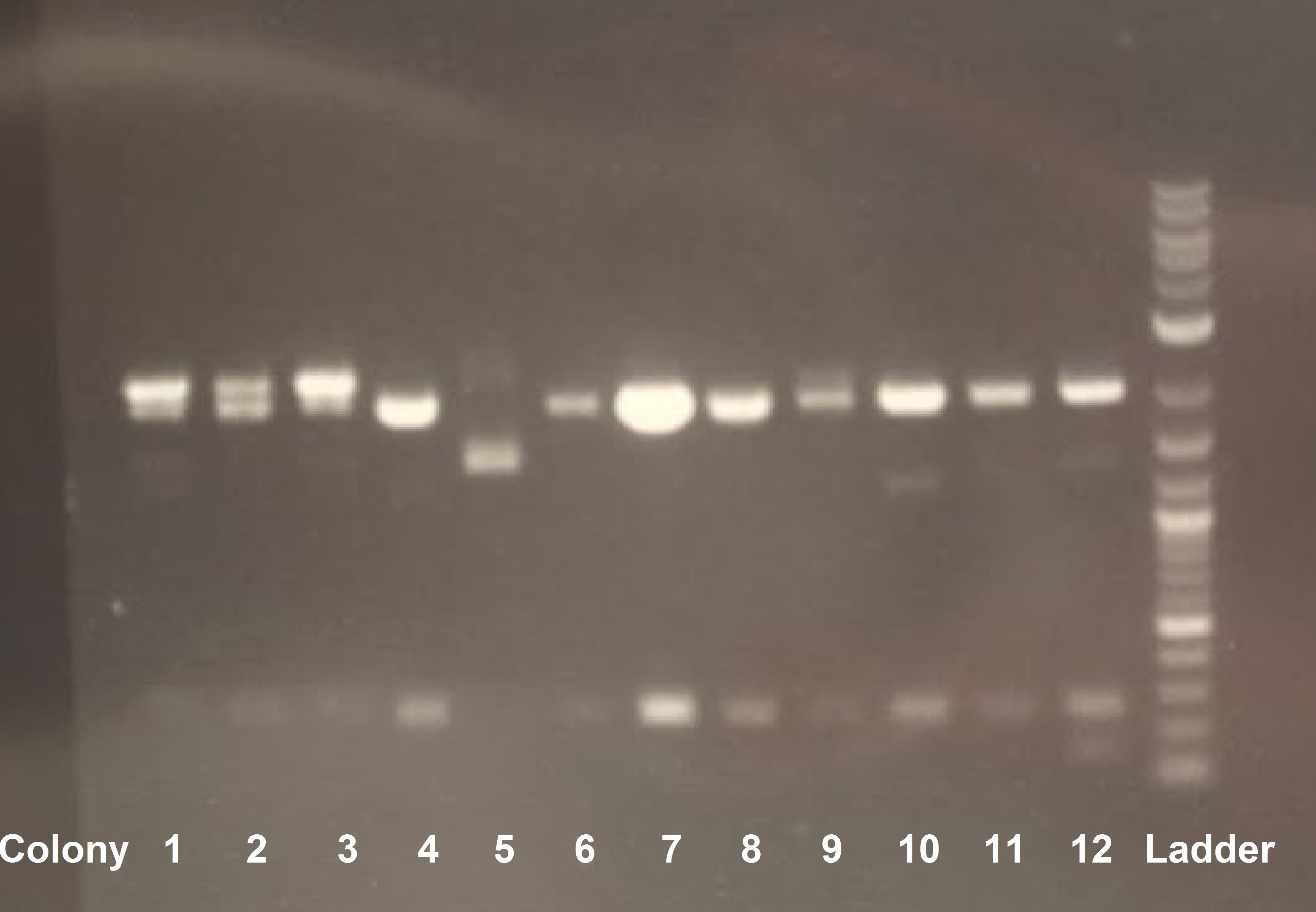
Due to an error in labeling the plate and miniprep tubes, we decided to redo the gel. Based on the lanes with the least amount of noise (colonies 7 and 8), we transformed our competent cells using the DNA corresponding to those colonies. An overnight will be done tomorrow for the experiment on the following day.
August 19th
We ran an inoculation experiment using the cells with the CUP1 codon-optimized gene. After realizing that the copper solution was interfering with our OD data, we decided to resuspend the cells in LB and use LB as our blank to gather the OD data.
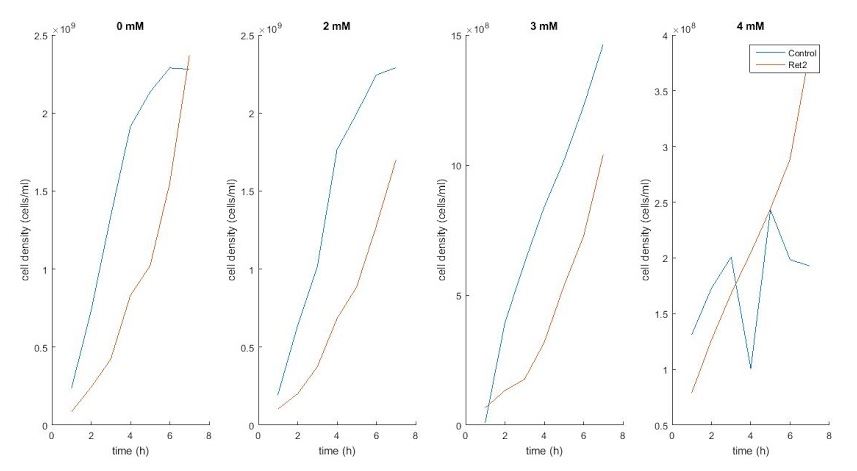
August 20th
We ran an SDS-PAGE gel using the samples collected from the experiment we did on August 19th. Since these were new gels, we made a few errors which resulted in unreadable results.
August 21th
We attempted the most recent SDS-PAGE gel again, with successful results. This gel showed a band in the Ret2 samples at a size of around 32 kDa, while our Metallothionein protein was expected to be around 6.5 kDa. We are pondering the implications of this data. We also ran two more gels with the experiment for Ret1 data.

August 25th
We ran an SDS-Gel on Ret1, which came out inconclusive.
September 13th
The LamB-Metallothionein fusion came, we assembled and transformed. Then, we ran our last inoculation experiment over a 10 hour time period.