Difference between revisions of "Team:Goettingen/Results"
| Line 270: | Line 270: | ||
<p><strong>RFP</strong></p> | <p><strong>RFP</strong></p> | ||
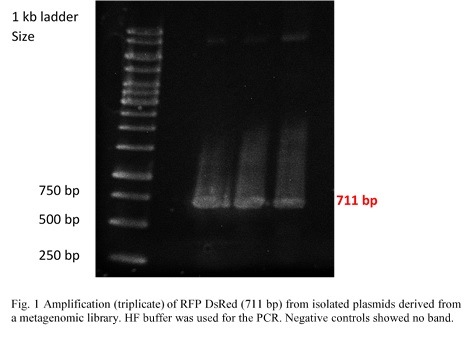
| − | <p>RFP (RFP DsRed) was amplified from pHT315_rfp by PCR (Fig.1). Colonies on a plate were given to us by the Applied and Genomic Microbiology department. Primers contained restriction sites for <em>Kpn</em>I and <em>Sac</em>I in order to make them compatible for insertion into the multiple cloning site of the pBAD/His A vector.</p> | + | <p>RFP (RFP DsRed) was amplified from pHT315_rfp by PCR (Fig.1). Colonies on a plate were given to us by the Applied and Genomic Microbiology department. Primers contained restriction sites for <em>Kpn</em>I and <em>Sac</em>I in order to make them compatible for insertion into the multiple cloning site of the pBAD/His A vector. The PCR products were examined in a 0,8% agarose gel by electrophoresis.</p> |
</html> | </html> | ||
[[File:PCR_RFP_DsRed_iGEM_Goettingen_2015.jpg|thumb|500px|center|<p style="text-align: justify;">Fig.1) A gel of the RFP DsRed PCR product showed the bands to be at the correct height.</p>]] | [[File:PCR_RFP_DsRed_iGEM_Goettingen_2015.jpg|thumb|500px|center|<p style="text-align: justify;">Fig.1) A gel of the RFP DsRed PCR product showed the bands to be at the correct height.</p>]] | ||
| Line 278: | Line 278: | ||
<p>After purification of the PCR product it was ligated into pJET1.2 by subcloning (blunt end ligation). This vector serves to clone the fluorescent protein without triggering its activity, which may interact with the expression vector or the chosen <em>E.coli </em>strain.</p> | <p>After purification of the PCR product it was ligated into pJET1.2 by subcloning (blunt end ligation). This vector serves to clone the fluorescent protein without triggering its activity, which may interact with the expression vector or the chosen <em>E.coli </em>strain.</p> | ||
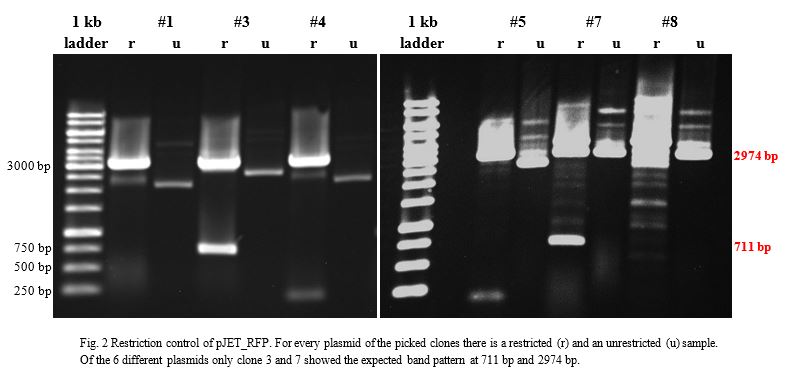
| − | <p>Ligation into pJET1.2 was followed according to the protocol in the methods collection. After over-night incubation colonies were picked, plasmids extracted with the QIAGEN QIAprep Spin Miniprep Kit | + | <p>Ligation into pJET1.2 was followed according to the protocol in the methods collection. After over-night incubation colonies were picked, plasmids extracted with the QIAGEN QIAprep Spin Miniprep Kit, restricted with <em>Kpn</em>I and <em>Sac</em>I and examined in a gel (Fig.2).</p> |
</html> | </html> | ||
| Line 287: | Line 287: | ||
<p>The next step was a big restriction of the whole plasmid extract with the correspondent pair of restriction enzymes. That allowed us to isolate the RFP inserts carrying the desired restriction sites (<em>Kpn</em>I and <em>Sac</em>I). The fragments were purified with the PEQLAB peqGOLD Gel Extraction Kit before T4 ligation. The RFP_3 insert was ligated into pBAD/His A by the T4 ligation system (sticky end ligation) according to the protocol in the methods collection and transformed into <em>E.coli</em> TOP10. This vector serves to express the fluorescent protein by triggering its activity.</p> After over-night incubation colonies were picked, plasmids extracted with the QIAGEN QIAprep Spin Miniprep Kit and restricted with <em>Kpn</em>I and <em>Sac</em>I.</p> | <p>The next step was a big restriction of the whole plasmid extract with the correspondent pair of restriction enzymes. That allowed us to isolate the RFP inserts carrying the desired restriction sites (<em>Kpn</em>I and <em>Sac</em>I). The fragments were purified with the PEQLAB peqGOLD Gel Extraction Kit before T4 ligation. The RFP_3 insert was ligated into pBAD/His A by the T4 ligation system (sticky end ligation) according to the protocol in the methods collection and transformed into <em>E.coli</em> TOP10. This vector serves to express the fluorescent protein by triggering its activity.</p> After over-night incubation colonies were picked, plasmids extracted with the QIAGEN QIAprep Spin Miniprep Kit and restricted with <em>Kpn</em>I and <em>Sac</em>I.</p> | ||
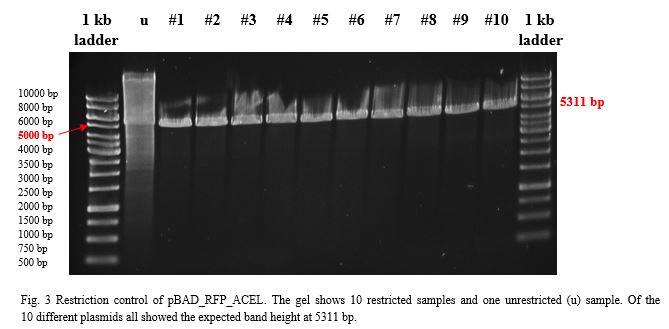
| − | <p>To build a component for our Flexosome, we decided to fuse pBAD_RFP with the ACEL (<em>Acetivibrio cellulolyticus</em>) dockerin. Both components, restricted with <em>Kpn</em>I and <em>Sac</em>I and purified with the PEQLAB peqGOLD Gel Extraction Kit were ligated by the T4 ligation system. After transformation into <em>E.coli</em> TOP10 and over-night incubation colonies were picked, plasmids extracted with the QIAGEN QIAprep Spin Miniprep Kit | + | <p>To build a component for our Flexosome, we decided to fuse pBAD_RFP with the ACEL (<em>Acetivibrio cellulolyticus</em>) dockerin. Both components, restricted with <em>Kpn</em>I and <em>Sac</em>I and purified with the PEQLAB peqGOLD Gel Extraction Kit were ligated by the T4 ligation system. After transformation into <em>E.coli</em> TOP10 and over-night incubation colonies were picked, plasmids extracted with the QIAGEN QIAprep Spin Miniprep Kit, restricted with <em>Eco</em>RI and examined in a gel (Fig.3).</p> |
</html> | </html> | ||
| Line 299: | Line 299: | ||
<p> </p> | <p> </p> | ||
<p><strong>MICROSCOPY</strong></p> | <p><strong>MICROSCOPY</strong></p> | ||
| − | <p><span lang="EN-US">To check the activity of RFP | + | <p><span lang="EN-US">To check the activity of RFP, an induction series with 0,2%, 2% and 5% L-Arabinose in the cell culture medium was prepared. Fluorescence microscopy was performed with the RFP DsRed filter (excitation at 536 nm, emission at 582 nm). Unfortunately no fluorescence could be detected throughout the whole project with different constructs. Neither in the pJET_RFP_3, nor in the pBAD constructs (pBAD_RFP_3 and pBAD_RFP_ACEL) induced with 0,2%, 2% and 5% L-Arabinose in the medium. Nevertheless every time the DNA sequences were correct (Sanger sequencing). It might be a problem of expression. |
<p> </p> | <p> </p> | ||
<p><strong>FUTURE PLANS</strong></p>In future we want to exchange our vector system (pET100 instead of pBAD) to ensure that pBAD is not somehow interfering with our chosen <em>E. coli</em> strain.</span></p> | <p><strong>FUTURE PLANS</strong></p>In future we want to exchange our vector system (pET100 instead of pBAD) to ensure that pBAD is not somehow interfering with our chosen <em>E. coli</em> strain.</span></p> | ||
Revision as of 17:59, 16 September 2015
Project Results
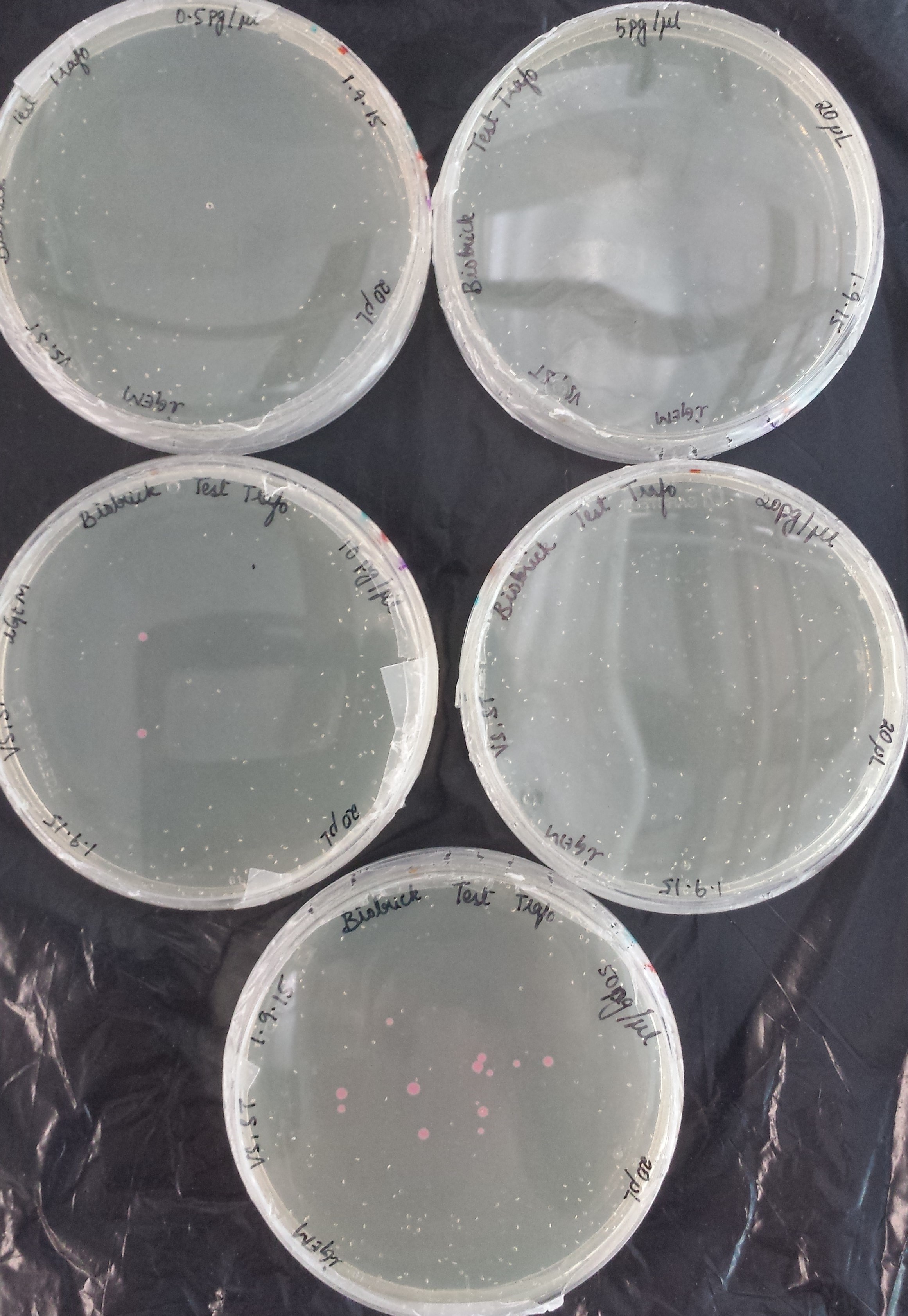
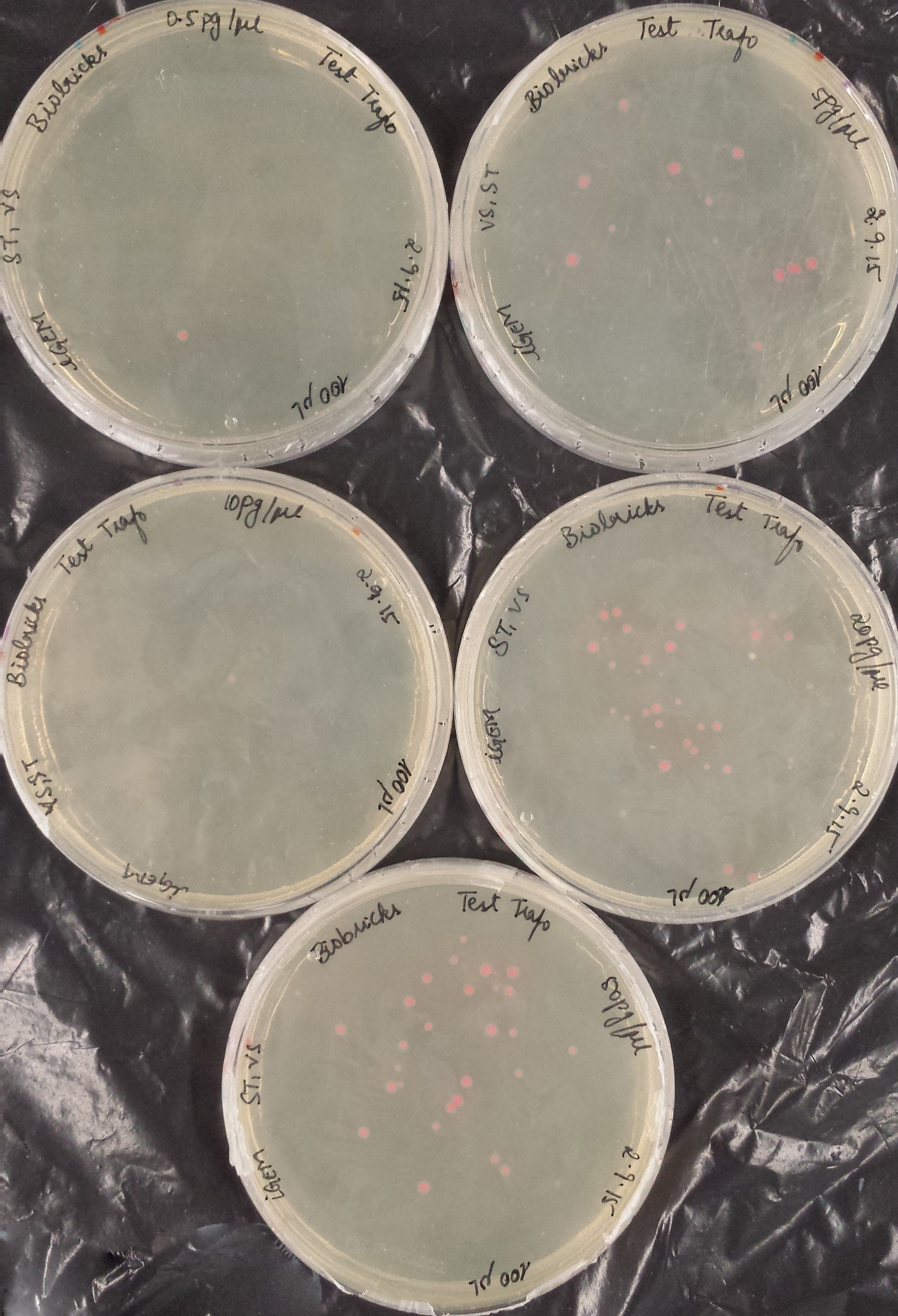
Transformation Efficiency Kit, RFP construct (iGEM)
RFP
Esterase and Phosphatase
Project Achievements
What should this page contain?
Here you can describe the results of your project and your future plans.
- Clearly and objectively describe the results of your work.
- Future plans for the project
- Considerations for replicating the experiments
You can also include a list of bullet points (and links) of the successes and failures you have had over your summer. It is a quick reference page for the judges to see what you achieved during your summer.
- A list of linked bullet points of the successful results during your project
- A list of linked bullet points of the unsuccessful results during your project. This is about being scientifically honest. If you worked on an area for a long time with no success, tell us so we know where you put your effort.
Inspiration
See how other teams presented their results.