Difference between revisions of "Team:Goettingen/Experiments"
JSchwanbeck (Talk | contribs) |
JSchwanbeck (Talk | contribs) |
||
| Line 130: | Line 130: | ||
<td valign="top" width="307"> | <td valign="top" width="307"> | ||
<p> | <p> | ||
| − | KH<sub>2</sub>PO<sub>4</sub> (MW: 136 | + | KH<sub>2</sub>PO<sub>4</sub> (MW: 136.09) |
</p> | </p> | ||
</td> | </td> | ||
<td valign="top" width="97"> | <td valign="top" width="97"> | ||
<p> | <p> | ||
| − | 7 | + | 7.0g |
</p> | </p> | ||
</td> | </td> | ||
| Line 147: | Line 147: | ||
<td valign="top" width="97"> | <td valign="top" width="97"> | ||
<p> | <p> | ||
| − | 15 | + | 15.14g |
</p> | </p> | ||
</td> | </td> | ||
| Line 166: | Line 166: | ||
</table> | </table> | ||
<p> | <p> | ||
| − | pH 6 | + | pH 6.8 with NaOH (ca. 10mL 1M NaOH) |
</p> | </p> | ||
<p> | <p> | ||
| Line 193: | Line 193: | ||
<td valign="top" width="97"> | <td valign="top" width="97"> | ||
<p> | <p> | ||
| − | 7 | + | 7.5mL |
</p> | </p> | ||
</td> | </td> | ||
| Line 208: | Line 208: | ||
<p> </p> | <p> </p> | ||
<p><span style="text-decoration: underline;">A: Stock reagents</span></p> | <p><span style="text-decoration: underline;">A: Stock reagents</span></p> | ||
| − | <p>1M IPTG (4 | + | <p>1M IPTG (4.76 g in 20ml Millipore-H2O) filter with blue 0.22 µm sterile filter and freeze at -20˚C</p> |
| − | <p>50mg/ml Kanamycin in Millipore H2O, filter with blue 0 | + | <p>50mg/ml Kanamycin in Millipore H2O, filter with blue 0.22 µm sterile filter and freeze at -20˚C</p> |
| − | <p>100mg/ml Ampicillin in 50% ethanol, filter with blue 0 | + | <p>100mg/ml Ampicillin in 50% ethanol, filter with blue 0.22 µm sterile filter and freeze at -20˚C</p> |
| − | <p>25mg/ml BCIP (in DMF) filter with blue 0 | + | <p>25mg/ml BCIP (in DMF) filter with blue 0.22 µm sterile filter and keep in a falcon tube wrapped with aluminum foil at 4˚C. BCIP: Biomol Nr. 2291 (MG 433.64)</p> |
<p>Glycerol (99%), autoclaved and kept at room temperature</p> | <p>Glycerol (99%), autoclaved and kept at room temperature</p> | ||
<p> </p> | <p> </p> | ||
<p><span style="text-decoration: underline;">B: Sperber medium</span></p> | <p><span style="text-decoration: underline;">B: Sperber medium</span></p> | ||
<p>1g Yeast extract</p> | <p>1g Yeast extract</p> | ||
| − | <p>3 | + | <p>3.5 ml 50% w/w Phytic acid</p> |
| − | <p>0 | + | <p>0.2g CaCl2</p> |
| − | <p>0 | + | <p>0.5g MgSO4</p> |
| − | <p>Adjust the pH to 7 | + | <p>Adjust the pH to 7.2 with NaOH</p> |
<p>Ad 2L of Millipore H2O</p> | <p>Ad 2L of Millipore H2O</p> | ||
<p>32g agar</p> | <p>32g agar</p> | ||
<p>Autoclave</p> | <p>Autoclave</p> | ||
<p> </p> | <p> </p> | ||
| − | <p>After autoclaving the medium must cool down to ca. | + | <p>After autoclaving the medium must cool down to ca. 50 ˚C. Now glycerol, IPTG, BCIP and the respective antiobiotic can be added.</p> |
<p> </p> | <p> </p> | ||
<p>For 2 L medium:</p> | <p>For 2 L medium:</p> | ||
| Line 240: | Line 240: | ||
<p style="text-align: center;"><span style="font-size: x-large; color: #888888;"><strong><span lang="EN-GB"></span></strong></span></p> | <p style="text-align: center;"><span style="font-size: x-large; color: #888888;"><strong><span lang="EN-GB"></span></strong></span></p> | ||
<p><span lang="EN-GB"> </span></p> | <p><span lang="EN-GB"> </span></p> | ||
| − | <p><span lang="EN-GB">To 500ml of LB Media add 7 | + | <p><span lang="EN-GB">To 500ml of LB Media add 7.5g of Agar and 5ml of Tributyrin and homogenize with a mixer.</span></p> |
<p><span lang="EN-GB">This culture medium must be directly sterilized by autoclaving at 121˚C for 20 min.</span></p> | <p><span lang="EN-GB">This culture medium must be directly sterilized by autoclaving at 121˚C for 20 min.</span></p> | ||
<p><span lang="EN-GB">If you wait too long it will be inhomogeneous again!</span></p> | <p><span lang="EN-GB">If you wait too long it will be inhomogeneous again!</span></p> | ||
| Line 310: | Line 310: | ||
<p>The stock solution will be stored at +4 °C.</p> | <p>The stock solution will be stored at +4 °C.</p> | ||
<p><strong>C: Prepare the agar plates</strong></p> | <p><strong>C: Prepare the agar plates</strong></p> | ||
| − | <p>When the medium cools down to around 60°C, put the bottle containing LB-agar base on a magnetic mixer. At the same time, disperse the substrate by flip the falcon tube several times. Then, pour the substrate into the LB-agar base gently until the substrate suspend evenly in the agar medium (For how much substrate should be poured, ask the supervisor). Other reagents like antibiotic can also be added. Ca. 3-5 ml substrate per Liter medium.</p> | + | <p>When the medium cools down to around 60 °C, put the bottle containing LB-agar base on a magnetic mixer. At the same time, disperse the substrate by flip the falcon tube several times. Then, pour the substrate into the LB-agar base gently until the substrate suspend evenly in the agar medium (For how much substrate should be poured, ask the supervisor). Other reagents like antibiotic can also be added. Ca. 3-5 ml substrate per Liter medium.</p> |
<p><strong>D: Preparation of agar plates contacting cellulose substrate</strong></p> | <p><strong>D: Preparation of agar plates contacting cellulose substrate</strong></p> | ||
<p>After dispersing substrate and adding antibiotic to LB-agar , pour the medium quickly with continuous stirring onto the plates and make sure to make a thin layer of medium. Let the plates to dry and store them at 4 °C.</p> | <p>After dispersing substrate and adding antibiotic to LB-agar , pour the medium quickly with continuous stirring onto the plates and make sure to make a thin layer of medium. Let the plates to dry and store them at 4 °C.</p> | ||
| Line 372: | Line 372: | ||
<ul> | <ul> | ||
<li>Add ethanol (96–100%) to Buffer PE before use.</li> | <li>Add ethanol (96–100%) to Buffer PE before use.</li> | ||
| − | <li>All centrifugation steps are carried out at | + | <li>All centrifugation steps are carried out at 17900 g in a conventional table-top microcentrifuge at room temperature.</li> |
</ul> | </ul> | ||
<p> </p> | <p> </p> | ||
| Line 388: | Line 388: | ||
<p>- Fractionate DNA fragments by running a agarose gel (Do not stain the gel with Ethidium bromide or expose the DNA to UV for too long).</p> | <p>- Fractionate DNA fragments by running a agarose gel (Do not stain the gel with Ethidium bromide or expose the DNA to UV for too long).</p> | ||
<p>- Add equal volume of Binding Buffer to the gel slice and incubate at 65ᵒC for 8 min. Vortex or mix every 2 to 3 min until the agarose dissolves completely. (0.2 g of gel equivalent to 0.2 ml)</p> | <p>- Add equal volume of Binding Buffer to the gel slice and incubate at 65ᵒC for 8 min. Vortex or mix every 2 to 3 min until the agarose dissolves completely. (0.2 g of gel equivalent to 0.2 ml)</p> | ||
| − | <p>- Pour the mixture into PerfectBind DNA column (which is placed in a 2 ml collection tube) and centrifuge for 1 min at | + | <p>- Pour the mixture into PerfectBind DNA column (which is placed in a 2 ml collection tube) and centrifuge for 1 min at 10000 x g.(max. 750 µl) Discard the flow-through and place the PerfectBind DNA column in the same tube. Repeat the steps if required.</p> |
| − | <p>- Add 300 µl of binding Buffer to the PerfectBind DNA column for the washing the contaminants and centrifuge for 1 min at | + | <p>- Add 300 µl of binding Buffer to the PerfectBind DNA column for the washing the contaminants and centrifuge for 1 min at 10000 x g. Discard the flow-through and place the column in the same tube</p> |
| − | <p>- Add 750 µl of CG Wash Buffer to the PerfectBind DNA column for the wash, incubate for 2 to 3 min and centrifuge for 1 min at | + | <p>- Add 750 µl of CG Wash Buffer to the PerfectBind DNA column for the wash, incubate for 2 to 3 min and centrifuge for 1 min at 10000 x g. Discard the flow-through and place the column in the same tube. Repeat this step once more.</p> |
| − | <p>- Centrifuge the PerfectBind DNA column once more in the 2 ml collection tube for 1 min at | + | <p>- Centrifuge the PerfectBind DNA column once more in the 2 ml collection tube for 1 min at 10000 x g to remove the residual wash buffer.</p> |
<p>- Place the PerfectBind DNA column in a clean 1.5 ml eppendorf tube.</p> | <p>- Place the PerfectBind DNA column in a clean 1.5 ml eppendorf tube.</p> | ||
| − | <p>- Add 50 µl of pre-warmed sterile water to the PerfectBind DNA column and incubate for 2 to 3 min (normally in a 2 step process of 30 µl in first elution step and 20 µl in second elution step) and centrifuge at | + | <p>- Add 50 µl of pre-warmed sterile water to the PerfectBind DNA column and incubate for 2 to 3 min (normally in a 2 step process of 30 µl in first elution step and 20 µl in second elution step) and centrifuge at 5000 x g for 1 min.</p> |
<p>- Store the purified DNA at -20ᵒC.</p> | <p>- Store the purified DNA at -20ᵒC.</p> | ||
<p> </p> | <p> </p> | ||
| Line 669: | Line 669: | ||
<li>Add the provided RNase A solution to buffer P1, mix, and store at 4 <sup>o</sup>C.</li> | <li>Add the provided RNase A solution to buffer P1, mix, and store at 4 <sup>o</sup>C.</li> | ||
<li>Add ethanol (96–100%) to Buffer PE before use.</li> | <li>Add ethanol (96–100%) to Buffer PE before use.</li> | ||
| − | <li>All centrifugation steps are carried out at | + | <li>All centrifugation steps are carried out at 17900 x g (13000 rpm) in a conventional table-top microcentrifuge at room temperature.</li> |
</ul> | </ul> | ||
<p> </p> | <p> </p> | ||
| Line 684: | Line 684: | ||
<p>- Add 350 μl Buffer N3 and mix immediately and thoroughly by inverting the tube 4–6 times.</p> | <p>- Add 350 μl Buffer N3 and mix immediately and thoroughly by inverting the tube 4–6 times.</p> | ||
| − | <p>- Centrifuge for 10 min at | + | <p>- Centrifuge for 10 min at 17900 x g in a table-top microcentrifuge.</p> |
<p>- Apply the supernatant to the QIAprep spin column by decanting. Centrifuge 60 s. Discard the flow-through.</p> | <p>- Apply the supernatant to the QIAprep spin column by decanting. Centrifuge 60 s. Discard the flow-through.</p> | ||
| Line 815: | Line 815: | ||
<p> </p> | <p> </p> | ||
<p><strong>Protocol as distributed by iGEM (modified) </strong></p> | <p><strong>Protocol as distributed by iGEM (modified) </strong></p> | ||
| − | <p>Spin down the DNA tubes from the Transformation Efficiency Kit to collect all of the DNA into the bottom of each tube prior to use. A quick spin of 20-30 seconds at | + | <p>Spin down the DNA tubes from the Transformation Efficiency Kit to collect all of the DNA into the bottom of each tube prior to use. A quick spin of 20-30 seconds at 8000-10000 rpm will be sufficient. Note: There should be 50 µL of DNA in each tube sent in the Kit.</p> |
<p>Thaw competent cells on ice. Label one 2.0 ml microcentrifuge tube for each concentration and then pre-chill by placing the tubes on ice.</p> | <p>Thaw competent cells on ice. Label one 2.0 ml microcentrifuge tube for each concentration and then pre-chill by placing the tubes on ice.</p> | ||
<p>Pipet 1 µL of DNA into each microcentrifuge tube. For each concentration, use a separate tube.</p> | <p>Pipet 1 µL of DNA into each microcentrifuge tube. For each concentration, use a separate tube.</p> | ||
| Line 979: | Line 979: | ||
<p><strong>Materials:</strong></p> | <p><strong>Materials:</strong></p> | ||
<ul> | <ul> | ||
| − | <li>Freshly prepared AAM Solution (Acetone, 5N H<sub>2</sub>SO<sub>4</sub>, | + | <li>Freshly prepared AAM Solution (Acetone, 5N H<sub>2</sub>SO<sub>4</sub>, 10 mM Ammonium molybdate, 2:1:1 v/v)</li> |
<li>50 mM Sodium acetate buffer, pH 5.0</li> | <li>50 mM Sodium acetate buffer, pH 5.0</li> | ||
<li>100 mM Pyrophosphate</li> | <li>100 mM Pyrophosphate</li> | ||
| Line 1,133: | Line 1,133: | ||
<p></p> | <p></p> | ||
<ul> | <ul> | ||
| − | <li>For 10 µl reaction add 1 µl of 10x Tango buffer , for 20 µl reaction add 2 µl of 10x Tango buffer and vice versa.</li> | + | <li>For 10 µl reaction add 1 µl of 10x Tango buffer, for 20 µl reaction add 2 µl of 10x Tango buffer and vice versa.</li> |
<li>Incubate at 37 <sup>o</sup>C for 2 hours.</li> | <li>Incubate at 37 <sup>o</sup>C for 2 hours.</li> | ||
<li>For inactivation incubation at 80°C for 5min. </li> | <li>For inactivation incubation at 80°C for 5min. </li> | ||
| Line 1,564: | Line 1,564: | ||
</td> | </td> | ||
<td valign="top" width="57"> | <td valign="top" width="57"> | ||
| − | <p>0 | + | <p>0.5 µl</p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 1,588: | Line 1,588: | ||
</td> | </td> | ||
<td valign="top" width="57"> | <td valign="top" width="57"> | ||
| − | <p>1 | + | <p>1.5 µl</p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 1,597: | Line 1,597: | ||
<p><span style="text-decoration: underline;"> </span></p> | <p><span style="text-decoration: underline;"> </span></p> | ||
<p>- <span style="text-decoration: underline;">Program: </span></p> | <p>- <span style="text-decoration: underline;">Program: </span></p> | ||
| − | <p>Initial denaturalization: 98˚C, | + | <p>Initial denaturalization: 98˚C, 5 min</p> |
| − | <p>Denaturalization: 96˚C, 45 | + | <p>Denaturalization: 96˚C, 45 s</p> |
| − | <p>Annealing: 70˚C, 30 | + | <p>Annealing: 70˚C, 30 s</p> |
| − | <p>Elongation: 72˚C, 25 | + | <p>Elongation: 72˚C, 25 s</p> |
<p></p> | <p></p> | ||
<p> </p> | <p> </p> | ||
| Line 2,103: | Line 2,103: | ||
<ul> | <ul> | ||
<p><strong>RFP microscopy</strong></p> | <p><strong>RFP microscopy</strong></p> | ||
| − | <p>To check if the transformed <em>E.coli</em> TOP10 show red fluorescence, the culture was examined by fluorescence microscopy. The RFP DsRed filter was used (excitation at 536 nm, emission at 582 nm). To prevent the cells from floating around they are fixed in 0 | + | <p>To check if the transformed <em>E.coli</em> TOP10 show red fluorescence, the culture was examined by fluorescence microscopy. The RFP DsRed filter was used (excitation at 536 nm, emission at 582 nm). To prevent the cells from floating around they are fixed in 0.8 %.</p> |
<p> </p> | <p> </p> | ||
<p><strong>Preparation of slides</strong></p> | <p><strong>Preparation of slides</strong></p> | ||
<ul> | <ul> | ||
| − | <li>Prepare 0 | + | <li>Prepare 0.8 % agarose with water and boil it up.</li> |
<li>Pipet 500 μL onto a slide and press another one on top. Let the agarose cure between both slides.</li> | <li>Pipet 500 μL onto a slide and press another one on top. Let the agarose cure between both slides.</li> | ||
<li>Carefully remove the upper slide and put the sample name in one corner the one with the agarose.</li> | <li>Carefully remove the upper slide and put the sample name in one corner the one with the agarose.</li> | ||
Revision as of 17:09, 17 September 2015
Media/Buffer
LB Medium
"Fat" LB Medium
Phosphatase Activity plates, Sperber media
Esterase Activity plates, with 1% Tributyrin
Cellulase activity plates
1x TAE Buffer
Cloning Methods
PCR product purification using QIAquick® PCR Purification Kit (QIAGEN)
PCR Gel extraction, peqGOLD Gel Extraction Kit
Blunt End Ligation in pJET1.2 vector –Clone JET PCR Cloning Kit– (Thermo Scientific)
Sticky End T4 Ligation (Thermo Scientific)
TOPO® Cloning protocol usingChampion™ pET Directional TOPO® Expression Kits (Thermo Fisher Scientific)
Plasmid transformation into chemically competent E. coli
Electroporation of BL21 cells with pJET_RFP
Plasmid Extraction - using QIAprep Spin Miniprep Kit (QIAGEN)
Plasmid Extraction - using peqGOLD Plasmid Miniprep Kit I (PEQLAB Technologies)
Competent Cells
Preparation of competent E.coli cells
Transformation Efficiency Kit, RFP construct (iGEM)
Protein Extraction and Purification
Protein Extraction (French Press) and Purification (Protino® Ni-IDA 2000 His-Tag protein purification, Macherey-Nagel)
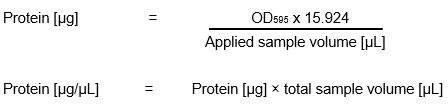
Bradford Assay
Activity Screens
Esterase activity test
Phosphatase activity test
Cellulase activity screening
Restriction Controls
Aan I (Psi I ) - thermo fisher scientific - restriction control protocol
Double digestion restriction control
Restriction control using fast and slow digestion enzymes
Scafoldin Restriction control
Esterase Restriction Control
Phosphatase Restriction Control
PCR Preparation Methods
Colony PCR
Phusion PCR
Sequencing
Protocol for Sanger sequencing
Overnight Sanger Sequencing
Fluorescence Microscopy
RFP microscopy