Difference between revisions of "Team:Sydney Australia/TransOpt"
| (44 intermediate revisions by the same user not shown) | |||
| Line 49: | Line 49: | ||
As experiments have shown that translation rate (and other affects discussed above) can have a substantial impact on protein expression, it is desirous that synthetic biology develop optimisation tools to account for these deciding influences. Based on the assumption that a gene is expressed relatively efficiently in its native host, optimisation approaches therefore aim to maintain translational kinetics under the different cellular conditions of the heterologous host. Though several fairly advanced tools for codon optimisation exist <sup>[https://2015.igem.org/Team:Sydney_Australia/TransOpt#References 1]</sup> many authors continue to employ simple rate-maximisation or harmonisation tools when transferring a gene between hosts. | As experiments have shown that translation rate (and other affects discussed above) can have a substantial impact on protein expression, it is desirous that synthetic biology develop optimisation tools to account for these deciding influences. Based on the assumption that a gene is expressed relatively efficiently in its native host, optimisation approaches therefore aim to maintain translational kinetics under the different cellular conditions of the heterologous host. Though several fairly advanced tools for codon optimisation exist <sup>[https://2015.igem.org/Team:Sydney_Australia/TransOpt#References 1]</sup> many authors continue to employ simple rate-maximisation or harmonisation tools when transferring a gene between hosts. | ||
| − | One common approach to optimisation involves selecting codons to “maximise” the translation rate, whether in terms of Codon Adaptation Index (CAI) or Codon Bias <sup>[https://2015.igem.org/Team:Sydney_Australia/TransOpt#References 1]</sup> in the hope of thereby maximising gene expression. However, this method frequently proves unreliable, with its failure very likely stemming from the need for slow-translation regions to allow proteins adequate time to fold. This has been experimentally demonstrated, where sequences in which fast-translating codons are selected result in substantially decreased protein activity.<sup>[https://2015.igem.org/Team:Sydney_Australia/TransOpt#References 8] </sup> | + | One common approach to optimisation involves selecting codons to “maximise” the translation rate, whether in terms of Codon Adaptation Index (CAI) or Codon Bias <sup>[https://2015.igem.org/Team:Sydney_Australia/TransOpt#References 1]</sup> in the hope of thereby maximising gene expression. However, this method frequently proves unreliable, with its failure very likely stemming from the need for slow-translation regions to allow proteins adequate time to fold. This has been experimentally demonstrated, and was confirmed by our own results (see [https://2015.igem.org/Team:Sydney_Australia/TransOpt#Experimental_Validation Experimental Validation]), where sequences in which fast-translating codons are selected result in substantially decreased protein activity.<sup>[https://2015.igem.org/Team:Sydney_Australia/TransOpt#References 8] </sup> |
Another popular method is Codon Harmonization <sup>[https://2015.igem.org/Team:Sydney_Australia/TransOpt#References 15]</sup> which (in its various forms) attempts to select codons in the heterologous expression host with usage frequencies (Codon Bias) matched as near as possible with those in the native expression host. This, however, is not necessarily the best method for aligning translation rates in different mRNA regions (as will be examined in following sections), nor does it generally consider secondary effects such as mRNA structure (e.g. as approximated by GC content) or tRNA abundances. | Another popular method is Codon Harmonization <sup>[https://2015.igem.org/Team:Sydney_Australia/TransOpt#References 15]</sup> which (in its various forms) attempts to select codons in the heterologous expression host with usage frequencies (Codon Bias) matched as near as possible with those in the native expression host. This, however, is not necessarily the best method for aligning translation rates in different mRNA regions (as will be examined in following sections), nor does it generally consider secondary effects such as mRNA structure (e.g. as approximated by GC content) or tRNA abundances. | ||
| Line 112: | Line 112: | ||
'''Algorithm 2''' | '''Algorithm 2''' | ||
| − | '''1.''' Input codon sequence (\(s_{in}\)), target rate (\(r_t\)) and GC Richness (\(GC_t\)) profiles, and averaging width (k, where \(k = 2*w + 1\) with w the averaging half-width as defined previously). Initialise offset i as \(i = 1\). | + | '''1.''' Input codon sequence (\(s_{in}\)), target rate (\(r_t\)) and GC Richness (\(GC_t\)) profiles, and averaging width (\(k\), where \(k = 2*w + 1\) with w the averaging half-width as defined previously). Initialise offset \(i\) as \(i = 1\). |
'''2.''' Randomly synonymously alter codons at locations \(i+nk\) where \(n=0,1,2...\). (so \(i,i+k,i+2k,...\)) in the initial sequence \((s_0)\) to form an altered sequence \((s_1)\). | '''2.''' Randomly synonymously alter codons at locations \(i+nk\) where \(n=0,1,2...\). (so \(i,i+k,i+2k,...\)) in the initial sequence \((s_0)\) to form an altered sequence \((s_1)\). | ||
| Line 118: | Line 118: | ||
'''3.''' Calculate new rate \((r_1)\) and GC Richness profiles \((GC_1)\) for altered codon sequence \((s_1)\). | '''3.''' Calculate new rate \((r_1)\) and GC Richness profiles \((GC_1)\) for altered codon sequence \((s_1)\). | ||
| − | '''4.''' Calculate the change in “energy” between the altered and original sequence at each location i+nk, given by: | + | '''4.''' Calculate the change in “energy” between the altered and original sequence at each location \(i+nk\), given by: |
$$\triangle E_{i+nk}=\sum^{i+nk+w}_{j=i+nk-w}{\alpha \times \left(|r_{0_j}-r_{t_j}|-|r_{1_j}-r_{t_j}|\right)+\beta \times \left(|GC_{0_j}-GC_{t_j}|-|GC_{1_j}-GC_{t_j}|\right)}+f$$ | $$\triangle E_{i+nk}=\sum^{i+nk+w}_{j=i+nk-w}{\alpha \times \left(|r_{0_j}-r_{t_j}|-|r_{1_j}-r_{t_j}|\right)+\beta \times \left(|GC_{0_j}-GC_{t_j}|-|GC_{1_j}-GC_{t_j}|\right)}+f$$ | ||
| − | Where \(alpha ,\ \beta\) are weighting factors for different terms in the optimisation function, in our case set to \( \alpha =1,\ \beta =0.3\), and f is an arbitrary function, in our case set as \(f = 0\). | + | Where \(\alpha ,\ \beta\) are weighting factors for different terms in the optimisation function, in our case set to \( \alpha =1,\ \beta =0.3\), and \(f\) is an arbitrary function, in our case set as \(f = 0\). |
| − | '''5.''' At each location \(i+nk\) in \(s_0\) accept the codon change in \(s_1\) if \(\triangle E_{i+nk} | + | '''5.''' At each location \(i+nk\) in \(s_0\) accept the codon change in \(s_1\) if \(\triangle E_{i+nk} \geq 0\) or if \(\triangle E_{i+nk}<0\) accept the change with probability: |
$$p\left(\triangle E_{i+nk}\right)=1-exp(\epsilon \times \triangle E_{i+nk})$$ | $$p\left(\triangle E_{i+nk}\right)=1-exp(\epsilon \times \triangle E_{i+nk})$$ | ||
| − | where \(ϵ\) is a scaling factor, in our case set so \(ϵ=1/k\). Otherwise, reject the changed codon (let the codon at location i+nk in \(s_0\) remain the same). | + | where \(ϵ\) is a scaling factor, in our case set so \(ϵ=1/k\). Otherwise, reject the changed codon (let the codon at location \(i+nk\) in \(s_0\) remain the same). |
| − | '''6.''' If \(i<k\) increment i by 1 \((i=i+1)\) and repeat from step 2 using the updated sequence \(s_0\). Otherwise, end algorithm and output sequence \(s_0\) and its corresponding rate (\(r_0\)) and GC Richness (\(GC_0\)) profiles. | + | '''6.''' If \(i<k\) increment \(i\) by 1 \((i=i+1)\) and repeat from step 2 using the updated sequence \(s_0\). Otherwise, end algorithm and output sequence \(s_0\) and its corresponding rate (\(r_0\)) and GC Richness (\(GC_0\)) profiles. |
---- | ---- | ||
[[File:USYDAlgorithm2.jpg | 520px | right | thumb | ''Figure 6'': A) Calculating the energy function used in optimisation, which is defined based on the blue shaded area. B) The step-wise optimization method which slowly moves along a sequence.]] | [[File:USYDAlgorithm2.jpg | 520px | right | thumb | ''Figure 6'': A) Calculating the energy function used in optimisation, which is defined based on the blue shaded area. B) The step-wise optimization method which slowly moves along a sequence.]] | ||
| − | Conceptually, Algorithm 2 makes changes to the sequence at discrete points separated such that a change at any one will not affect the rate or GC Richness (which are averaged within a width w of a given point) at another. Then an energy value is calculated, which is used to drive the heterologous rate and GC Richness profiles towards their target profiles. Mathematically, this is equivalent to minimising the area shaded blue in the ''Figure 6 A'' about each point. | + | Conceptually, Algorithm 2 makes changes to the sequence at discrete points separated such that a change at any one will not affect the rate or GC Richness (which are averaged within a width \(w\) of a given point) at another. Then an energy value is calculated, which is used to drive the heterologous rate and GC Richness profiles towards their target profiles. Mathematically, this is equivalent to minimising the area shaded blue in the ''Figure 6 A'' about each point. |
| − | This area is calculated in the energy equation as the sum of absolute deviations between the various profiles, and is compared between the sequence with randomly altered codons (\(s_1\)) and the initial sequence (\(s_0\)). A positive energy therefore corresponds to a segment of r_1 in which the shaded area is smaller in magnitude than the corresponding area about the same point in \(r_0\) (the unaltered sequence). Hence, by accepting changes that create a positive energy we are selecting sequences which reduce this absolute deviation between current and target profiles, thereby driving the heterologous profiles towards the target profiles. | + | This area is calculated in the energy equation as the sum of absolute deviations between the various profiles, and is compared between the sequence with randomly altered codons (\(s_1\)) and the initial sequence (\(s_0\)). A positive energy therefore corresponds to a segment of \(r_1\) in which the shaded area is smaller in magnitude than the corresponding area about the same point in \(r_0\) (the unaltered sequence). Hence, by accepting changes that create a positive energy we are selecting sequences which reduce this absolute deviation between current and target profiles, thereby driving the heterologous profiles towards the target profiles. |
This is repeated as we increment the offset (\(i\)) in step 6 and move through the sequence of codons, which is demonstrated for the first iteration in ''Figure 6 B''. | This is repeated as we increment the offset (\(i\)) in step 6 and move through the sequence of codons, which is demonstrated for the first iteration in ''Figure 6 B''. | ||
| Line 147: | Line 147: | ||
==Overview Flowchart== | ==Overview Flowchart== | ||
| − | Combining Algorithms 1 and 2 we can create a summarised flow chart (''Figure 7'') for our optimisation procedure. It has not escaped our notice that Algorithm 1 repeats indefinitely. By design the optimiser will continually generate and display better-optimised sequences, | + | Combining Algorithms 1 and 2 we can create a summarised flow chart (''Figure 7'') for our optimisation procedure. It has not escaped our notice that Algorithm 1 repeats indefinitely. By design the optimiser will continually generate and display better-optimised sequences, asking the user every 30 iterations whether they would like to terminate the process. |
[[File:USYDAlgorithm3.jpg | 550px | center | thumb | ''Figure 7'': Flow chart describing the overall optimisation process.]] | [[File:USYDAlgorithm3.jpg | 550px | center | thumb | ''Figure 7'': Flow chart describing the overall optimisation process.]] | ||
| Line 155: | Line 155: | ||
Running the TransOpt algorithm with an arbitrary mRNA (in this case, luciferase <sup>[https://2015.igem.org/Team:Sydney_Australia/TransOpt#References 8] </sup>), as done in ''Figure 8'', can be illustrative for demonstration of its capabilities. A number of observations are apparent: | Running the TransOpt algorithm with an arbitrary mRNA (in this case, luciferase <sup>[https://2015.igem.org/Team:Sydney_Australia/TransOpt#References 8] </sup>), as done in ''Figure 8'', can be illustrative for demonstration of its capabilities. A number of observations are apparent: | ||
| − | + | * The rate and GC Richness profiles for the TransOpt optimised sequence (yellow) match very closely with the target values (blue). This shows the algorithm is performing as desired: It is forcing both the rate profile and GC Richness profile to converge to their target profiles. | |
| − | + | * The rate profile for the sequence with standard harmonization (red) does not accurately match the target profile, and in many areas is worse than the wild type sequence expressed in the heterologous host without any optimisation whatsoever. | |
| − | + | * The GC content profile for the sequence with standard harmonization departs significantly from the target profile. The target profile would be achieved by expression in the heterologous host without adjustment, as the GC Richness is a function of the codon sequence, not organism. | |
| − | + | * The output from TransOpt typically converges to a reasonable result within ~5 iterations, doing so in approximately 10 seconds on a standard laptop computer. However, as the user is required to stop optimisation once they deem an acceptable solution has been reached, the algorithm can run indefinitely to gradually improve its results. | |
| − | [[File:USYDRes1.jpg | | + | |
| + | [[File:USYDRes1.jpg | 800px | center | thumb | ''Figure 8'': Typical TransOpt output, run with a 585 bp luciferase sequence (as in ''Figure 5'').]] | ||
| Line 169: | Line 170: | ||
---- | ---- | ||
| − | + | ==Methodology== | |
| − | In order to understand | + | In order to better understand how the algorithm performed in achieving its mandate, it was experimentally validated via measuring the fluorescence of variants of ''Bacillus subtilis'' fluorescent protein (BsFP), different at the transcriptome but not proteomics level. This was taken from its natural host ''B. subtilis'', and expressed in ''E. coli'', which meant utilising the tRNA GCNs for these two organisms for rate quantisation.<sup>[https://2015.igem.org/Team:Sydney_Australia/TransOpt#References 27]</sup> As in many other studies, the fluorescence of protein variants is assumed to be proportional to the level of protein expression and hence successful folding, which is what the algorithm aims to optimise. For more detailed explanation of the experimental design please refer to the [https://2015.igem.org/Team:Sydney_Australia/Results '''results page.'''] |
To assess the performance of a TransOpt variant of this protein we produced four synonymous strains by different methods: | To assess the performance of a TransOpt variant of this protein we produced four synonymous strains by different methods: | ||
# BsFP-WT: native sequence from the native host B.subtilis | # BsFP-WT: native sequence from the native host B.subtilis | ||
# BsFP-fast: all codons replaced with their synonyms possessing the highest rate, quantised using the approach of Spencer ''et al''<sup>[https://2015.igem.org/Team:Sydney_Australia/TransOpt#References 8] </sup> | # BsFP-fast: all codons replaced with their synonyms possessing the highest rate, quantised using the approach of Spencer ''et al''<sup>[https://2015.igem.org/Team:Sydney_Australia/TransOpt#References 8] </sup> | ||
| − | # BsFP-standard: harmonised BsFbFP generated via standard harmonisation using the rate quantisation approach of Spencer ''et al''<sup>[https://2015.igem.org/Team:Sydney_Australia/TransOpt#References 8] </sup> discussed above. | + | # BsFP-standard: harmonised BsFbFP generated via standard harmonisation (as proposed by Angov ''et al''<sup>[https://2015.igem.org/Team:Sydney_Australia/TransOpt#References 15] </sup>) using the rate quantisation approach of Spencer ''et al''<sup>[https://2015.igem.org/Team:Sydney_Australia/TransOpt#References 8] </sup> discussed above. |
| − | # BsFP-TransOpt: | + | # BsFP-TransOpt: optimised sequence generated using the TransOpt algorithm |
| + | |||
| + | |||
| + | |||
| + | [[File:ExpTO.jpg | 900px | center | thumb | ''Figure 9'': Rate and GC Richness profiles for the four sequence variants produced. Note that the WT Sequence in ''e coli'' has an identical GC Richness as when in llBacillus subtilis'', and hence it was not included in the lower plot. In both cases, the "target" rate profile is that for the WT Sequence in ''Bacillus subtilis''. ]] | ||
| + | |||
| + | In ''Figure 9'' the translation rate and GC Richness profiles for these four sequences are presented, and a number of features are evident: | ||
| + | |||
| + | *The rate profile of the WT sequence when expressed unaltered in ''e coli'' is not actually ''that'' far from the target profile (WT in ''Bacillus subtilis''). This implies that the difference between the two organisms, in terms of our quantisation for translation rate, is not particularly great. | ||
| + | |||
| + | *The Fast-optimised sequence has, as intended, substantially increased the translation rate at all locations in the sequence. In this case this has also resulted in an increased GC Richness throughout the sequence. | ||
| + | |||
| + | *The Standard harmonised sequence matches fairly well with the wild-type ''Bacillus subtilis'' target sequence in both profiles. Its rate profile matches more closely with the target sequence than the un-altered WT sequence when expressed in ''e coli'', however it is outperformed by the TransOpt sequence. | ||
| + | *The TransOpt sequence matches closely with the target rate profile, outperforming both the standard-optimised and WT sequence, and also matches well with the target GC Richness profile. | ||
| − | [[File:USYDfluro.jpg | 650px | center | thumb | ''Figure | + | ==Results== |
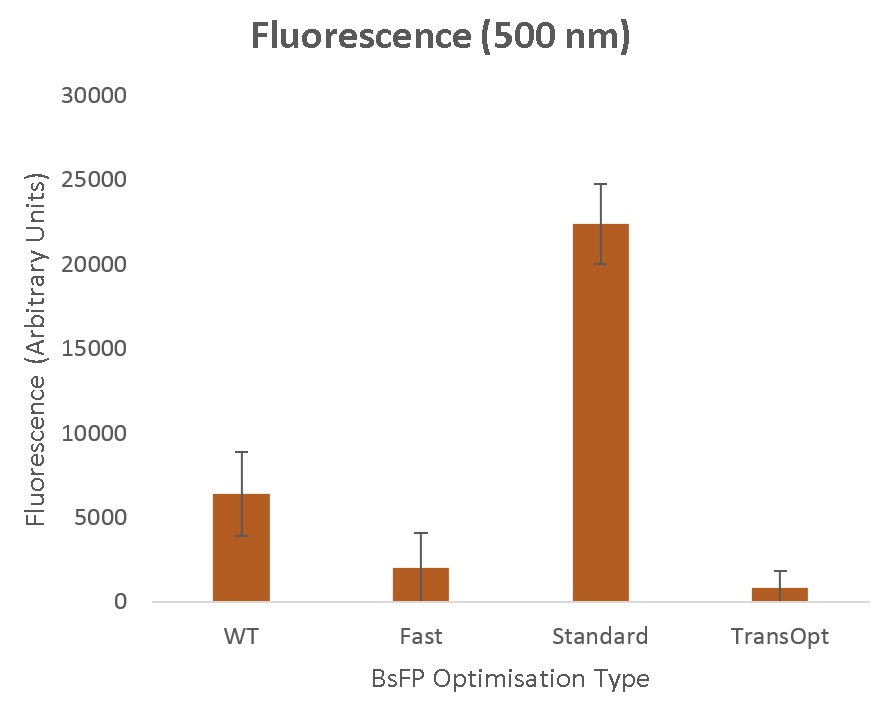
| + | Our experimental results, presented with more technical detail on the [https://2015.igem.org/Team:Sydney_Australia/Results Project Results] page, are summarized in ''Figure 10''. | ||
| + | [[File:USYDfluro.jpg | 650px | center | thumb | ''Figure 10'': Fluorescence measurements (in arbitrary units) for BsFP fluorescent protein. From left to right the strains are wild type (WT), fast codon optimised (fast), standard codon harmonisation (standard), TransOpt codon optimisation (TransOpt). ]] | ||
| − | |||
'''Standard Codon Harmonisation''' | '''Standard Codon Harmonisation''' | ||
| − | The sequence derived using standard codon harmonisation methods showed the strongest fluorescence, improving on the wild type sequence by a factor of more than | + | The sequence derived using standard codon harmonisation methods showed the strongest fluorescence, improving on the wild type sequence by a factor of more than 3. This result supported by previous experimental studies of fluorescence proteins, where it has been demonstrated that this commonly used harmonisation algorithm can substantially improve performance (for example, in the work of Angov ''et al'' <sup>[https://2015.igem.org/Team:Sydney_Australia/TransOpt#References 15]</sup>). To perform this harmonisation we used the translation rate quantisation methods of Spencer ''et al''<sup>[https://2015.igem.org/Team:Sydney_Australia/TransOpt#References 8] </sup>, and the strong expression here therefore provides evidence for the effectiveness of these methods. In ''Figure 11'' it is apparent that the relative utilisation of different codons in this variant is closest to that in the wild type. In ''Figure 9'' we see that, despite the weaknesses we believe exist in this optimisation method, in this simple protein it has provided fairly well-matching translation rate and GC Richness profiles. |
'''Fast Codon Utilisation ''' | '''Fast Codon Utilisation ''' | ||
| Line 192: | Line 207: | ||
The fast optimised sequence presented a substantially decreased fluorescence when compared to the wild type sequence. This is also as anticipated, and agrees with previous results which have shown the necessity for both slow- and fast-translating regions in mRNA.<sup>[https://2015.igem.org/Team:Sydney_Australia/TransOpt#References 14]</sup> In fact, our result aligns very closely with the work of Spencer ''et al'',<sup>[https://2015.igem.org/Team:Sydney_Australia/TransOpt#References 8] </sup> who similarly developed a synonymous sequence using the fastest-translating codons, and found its activity dropped to approximately half of its wild type performance. | The fast optimised sequence presented a substantially decreased fluorescence when compared to the wild type sequence. This is also as anticipated, and agrees with previous results which have shown the necessity for both slow- and fast-translating regions in mRNA.<sup>[https://2015.igem.org/Team:Sydney_Australia/TransOpt#References 14]</sup> In fact, our result aligns very closely with the work of Spencer ''et al'',<sup>[https://2015.igem.org/Team:Sydney_Australia/TransOpt#References 8] </sup> who similarly developed a synonymous sequence using the fastest-translating codons, and found its activity dropped to approximately half of its wild type performance. | ||
| − | A number of possible causes for this exists. For example, when using only fast-translating codons there may be insufficient time for co-translational folding of the protein, hampering its activity. However, another potential cause in this case (as examined qualitatively by our [https://2015.igem.org/Team:Sydney_Australia/Modeling/visualisation visualisation tools]) is tRNA starvation: If all codons that encode a single amino acid are substituted by for a single codon species, that species' utilisation throughout the sequence will be substantially increased, potentially resulting in starvation of its corresponding tRNA species during translation. | + | A number of possible causes for this exists. For example, when using only fast-translating codons there may be insufficient time for co-translational folding of the protein, hampering its activity. However, another potential cause in this case (as examined qualitatively by our [https://2015.igem.org/Team:Sydney_Australia/Modeling/visualisation visualisation tools]) is tRNA starvation: If all codons that encode a single amino acid are substituted by for a single codon species, that species' utilisation throughout the sequence will be substantially increased, potentially resulting in starvation of its corresponding tRNA species during translation.<sup>[https://2015.igem.org/Team:Sydney_Australia/TransOpt#References 31]</sup> This is apparent in ''Figure 11'', where it can be seen that the fast variant utilises a far smaller subset of the available codons, but uses each with a higher frequency. In the cell this might therefore be exhibited in starvation of these particular species, which is simulated by our [https://2015.igem.org/Team:Sydney_Australia/Modeling/visualisation visualisation tools]. Not only can this slow translation, but may in extreme cases result in stalling, which can activate ribosomal terminating mechanisms causing premature termination of translation.<sup>[https://2015.igem.org/Team:Sydney_Australia/TransOpt#References 32]</sup> Either of these mechanisms can cause an decrease in the cell's overall protein production, which would correspond to a decrease in fluorescence as our experimental results have demonstrated. |
'''TransOpt''' | '''TransOpt''' | ||
| − | The TransOpt optimised sequence showed the weakest fluorescence, performing | + | The TransOpt optimised sequence showed the weakest fluorescence, performing only marginally better than, and within experimental error of, the control measurement. This is not the result we had hoped for or expected, but as with all good science, sometimes important lessons can come from negative outcomes. |
| − | A number of potential causes for this failure exist, central among which might be our choice of test protein, BsFP. Though this comes from ''B. subtilis'', as we have demonstrated even in its wild type form it can function effectively in ''e. coli'', potentially due to its relative simplicity and short length. A major motivation behind TransOpt is the optimisation of sequences to account for both sections of fast- and slow-translating proteins, as well as mRNA folding. So, without a sufficiency complex protein that requires these controlling factors during translation, perhaps it is unsurprising that TransOpt would not outperform standard codon harmonisation methods. However, this does not explain why the fluorescence measured by TransOpt is so substantially decreased that it fails to (within error bars) outperform control measurements with non-fluorescent samples. One possible explanation may be that the TransOpt method has introduced a reliance on codon species that the heterologous expression host is unable to support. | + | A number of potential causes for this failure exist, central among which might be our choice of test protein, BsFP. Though this comes from ''B. subtilis'', as we have demonstrated even in its wild type form it can function effectively in ''e. coli'', potentially due to its relative simplicity and short length. A major motivation behind TransOpt is the optimisation of sequences to account for both sections of fast- and slow-translating proteins, as well as mRNA folding. So, without a sufficiency complex protein that requires these controlling factors during translation, perhaps it is unsurprising that TransOpt would not outperform standard codon harmonisation methods. This is illustrated by ''Figure 9'', where we see that both the WT and standard-optimised sequences do not differ very substantially from their target profiles, and hence we would expect them to perform ''relatively'' well. However, this does not explain why the fluorescence measured by TransOpt is so substantially decreased that it fails to (within error bars) outperform control measurements with non-fluorescent samples. One possible explanation may be that the TransOpt method has introduced a reliance on codon species that the heterologous expression host is unable to support. |
| − | We can identify some such species if we compare codon usage between the different protein variants. This reveals four codons (GCC, CTA, TCG and ACT), which are ''not'' used in either the WT, Fast or Standard variants, but ''are'' used in the TransOpt variant. Therefore, if our ''e. coli'' strain was unable to effectively translate any one of these codons, for whatever reason, this could cause a failure in protein production. A potential target for investigation would be the last of these codons, ACT, for which the corresponding tRNA (AGT) in ''e. coli'' has copy number zero, and it thus must rely on the wobble interaction with other species for translation. | + | We can identify some such species if we compare codon usage between the different protein variants. This reveals four codons (GCC, CTA, TCG and ACT, marked by red circles in ''Figure 11''), which are ''not'' used in either the WT, Fast or Standard variants, but ''are'' used in the TransOpt variant. Therefore, if our ''e. coli'' strain was unable to effectively translate any one of these codons, for whatever reason, this could cause a failure in protein production. A potential target for investigation would be the last of these codons, ACT, for which the corresponding tRNA (AGT) in ''e. coli'' has copy number zero, and it thus must rely on the wobble interaction with other species for translation. If we examine the overall codon usage for the TransOpt sequence (''Figure 11'') we see that it differs more substantially from the wild type than the standard harmonisation variant, though less so than the fast variant. We can also see numerous locations where the fast and standard optimised sequence utilise codons not found in the wild type sequence, however, this did not appear to prevent their expression. |
| + | |||
| + | [[File:CodonHistogram.jpg | 750px | center | thumb | ''Figure 11'': Histogram of usage frequency for different codons in the different BsFP variants. The red dots in the TransOpt histogram indicate codons used by TransOpt which are not used by any of the other three variants.]] | ||
=Possible Extensions= | =Possible Extensions= | ||
'''As exhibited by our experimental results,''' there appear to potentially be a number of shortcomings in our newly designed TransOpt approach. However, we believe that due to its scientific underpinning this method should not be discarded, and may be improved substantially by future work. | '''As exhibited by our experimental results,''' there appear to potentially be a number of shortcomings in our newly designed TransOpt approach. However, we believe that due to its scientific underpinning this method should not be discarded, and may be improved substantially by future work. | ||
| − | Though we believe that the failure of TransOpt to yield reliable function of BsFP may be a result of this protein's simplicity, short length, or potential errors in our fluorescence measurement experiments, the analysis of these results does highlight the possibility for tRNA starvation within the cell to play a role in protein expression. The mechanisms by which this occurs are demonstrated by our [https://2015.igem.org/Team:Sydney_Australia/Modeling/visualisation visualisation tools]. | + | Though we believe that the failure of TransOpt to yield reliable function of BsFP may be a result of this protein's simplicity, short length, or potential errors in our fluorescence measurement experiments, the analysis of these results does highlight the possibility for tRNA starvation within the cell to play a role in protein expression. The mechanisms by which this occurs are demonstrated by our [https://2015.igem.org/Team:Sydney_Australia/Modeling/visualisation visualisation tools]. Furthermore, for this particular protein the unchanged WT and standard-optimised sequences match quite well with the target profiles in ''Bacillus subtilis'' (''Figure 9''), leaving the TransOpt algorithm little room to make any substantial improvement. |
| − | A major improvement for TransOpt would therefore be inclusion of measures to account for this starvation: This would necessitate an additional parameter for optimisation | + | A major improvement for TransOpt would therefore be inclusion of measures to account for this starvation: This would necessitate an additional parameter for optimisation; an expression to account for the overall usage frequency of each codon (as a total for the entire mRNA sequence, as plotted in ''Figure 11''). Matching the ratios of these usages (as best as possible) to the tRNA GCNs would help to minimise the chance of tRNA starvation, as we would be utilising tRNAs at a rate as close as possible to the rate at which they are being produced. The astute reader will notice that in this we are proposing something similar to past efforts in codon optimisation: The matching of the codon usage between organisms. In the past this was done because codon usage was believed to correlate with rate of translation in some way. However, our motivation is substantially different: We hypothesize that codon usage instead balances with the differing rates of production between tRNA species, and in doing so regulates (and perhaps prevents) tRNA starvation. |
| − | '''Other possible extensions''' of TransOpt include | + | '''Other possible extensions''' of TransOpt include improvement of parameter estimations, and addition of other metrics during optimisation: |
| − | At present the TransOpt algorithm treats the rate and GC Richness profiles, which it aims to optimise, entirely independently of another. This leaves open the possibility for added optimisation terms in the energy equation, as represented by the arbitrary function f inserted at its end. Therefore, if in the future alternate measures of an mRNA’s “fitness” are derived, which are in some way a function of its codon sequence, the algorithm can be easily extended to optimise the profile of these measures: All that needs happen is their value be calculated and fed through the algorithm, much in the same way that the GC Richness value is at present. | + | At present the TransOpt algorithm treats the rate and GC Richness profiles, which it aims to optimise, entirely independently of another. This leaves open the possibility for added optimisation terms in the energy equation, as represented by the arbitrary function \(f\) inserted at its end. Therefore, if in the future alternate measures of an mRNA’s “fitness” are derived, which are in some way a function of its codon sequence, the algorithm can be easily extended to optimise the profile of these measures: All that needs happen is their value be calculated and fed through the algorithm, much in the same way that the GC Richness value is at present. |
| − | The weighting factors \((\alpha \ \&\ \beta )\) in the energy equation are used to determine the relative weighting of the rate and GC Richness optimisation terms. Such semi-arbitrary factors are ubiquitous in a range of optimisation techniques, as a means to scale the importance of different quantities to be optimised, and are generally (as they have been here) experimentally determined. Via greater experimentation with our algorithm it may be possible to choose better factors to replace these, which would certainly be necessary should we include any additional optimisation terms. This might serve to decrease convergence time of the algorithm. | + | The weighting factors \((\alpha \ \&\ \beta )\) in the energy equation are used to determine the relative weighting of the rate and GC Richness optimisation terms. Such semi-arbitrary factors are ubiquitous in a range of optimisation techniques, as a means to scale the importance of different quantities to be optimised, and are generally (as they have been here) experimentally determined. Via greater experimentation with our algorithm it may be possible to choose better factors to replace these, which would certainly be necessary should we include any additional optimisation terms. This might serve to decrease convergence time of the algorithm, however, we found similar resultant sequences were formed when these parameters were changed, so perhaps this is unlikely to improve the algorithm's performance from a biological perspective. |
Though we have used the quantisation of codon translation rate as proposed by Spencer ''et al'' <sup>[https://2015.igem.org/Team:Sydney_Australia/TransOpt#References 8] </sup> this choice makes no impact on the TransOpt algorithm’s mechanism. The rate quantisation is only responsible for providing data for different codon rates, which being essentially arbitrary to the algorithm’s function can be changed to anything desired. Therefore, it is possible to easily alter the TransOpt algorithm to utilise any quantisation of rate (for example, the Codon Usage Frequency / Codon Bias). That said, due to the strong fluorescence of our standard harmonised BsFP variant (which utilised the Spencer ''et al'' rate quantisation), it appears that the Spencer ''et al'' rate quantisation is functioning (at least to some extent) as intended. | Though we have used the quantisation of codon translation rate as proposed by Spencer ''et al'' <sup>[https://2015.igem.org/Team:Sydney_Australia/TransOpt#References 8] </sup> this choice makes no impact on the TransOpt algorithm’s mechanism. The rate quantisation is only responsible for providing data for different codon rates, which being essentially arbitrary to the algorithm’s function can be changed to anything desired. Therefore, it is possible to easily alter the TransOpt algorithm to utilise any quantisation of rate (for example, the Codon Usage Frequency / Codon Bias). That said, due to the strong fluorescence of our standard harmonised BsFP variant (which utilised the Spencer ''et al'' rate quantisation), it appears that the Spencer ''et al'' rate quantisation is functioning (at least to some extent) as intended. | ||
| − | Further study could also advise the choice of averaging width (\(w\)), which impacts the GC Richness and rate profiles. At present this is set to the value recommended by Spencer ''et al''<sup>[https://2015.igem.org/Team:Sydney_Australia/TransOpt#References 8] </sup> and is meant to reflect a characteristic length over which the translation process’s kinetics are impacted by the mRNA sequence. Decreasing this value would allow finer-resolution matching of a sequence to the target rate and GC Richness profiles, but would come at the expense of this match’s accuracy. To properly assess this parameter’s impact greater experimental data would be necessary, which could involve further study on the interdependency of translation rate and protein folding.<sup>[https://2015.igem.org/Team:Sydney_Australia/TransOpt#References 9] </sup> | + | Further study could also advise the choice of averaging width (\(w\)), which impacts the GC Richness and rate profiles. At present this is set to the value recommended by Spencer ''et al''<sup>[https://2015.igem.org/Team:Sydney_Australia/TransOpt#References 8] </sup> and is meant to reflect a characteristic length over which the translation process’s kinetics are impacted by the mRNA sequence. Decreasing this value would allow finer-resolution matching of a sequence to the target rate and GC Richness profiles, but would come at the expense of this match’s accuracy. If we set \(w=1\), then TransOpt would essentially return the same sequence as standard harmonisation tools: In each individual location it will return for the best matching rate. To properly assess this parameter’s impact greater experimental data would be necessary, which could involve further study on the interdependency of translation rate and protein folding.<sup>[https://2015.igem.org/Team:Sydney_Australia/TransOpt#References 9] </sup> |
| − | + | ||
| − | + | ||
| + | =Conclusion= | ||
| + | We have developed a new approach to codon optimisation, TransOpt, which aims to match several properties of a sequence in its heterologous expression host to that in the expression host. The mathematical approach to optimisation, simulated annealing, works exceptionally well, and demonstrates a capability to match these properties very accurately between sequences. However, experimental results demonstrated that in our test case a TransOpt optimized sequence did not function as expected: It resulted in negligible expression of a fluorescent protein. We identified a number of possible causes, such as this protein's simplicity and the fact that the TransOpt sequence utilised a number of codons not found in any of the other, more successful, variants. These variants included a sequence optimised by standard harmonisation methods, which showed a substantial increase in fluorescence over the wild type sequence, and a sequence in which only fast-translating codons were selected, which showed a substantial decrease in fluorescence compared to the wild type. These outcomes confirm previously published results regarding codon usage, and demonstrate that our rate quantisation metrics are functioning (to some degree) as intended. Finally, future improvements to the TransOpt algorithm are identified, and we suggest topics for future study which could more rigorously test certain features of TransOpt. | ||
| Line 288: | Line 305: | ||
<sup>30 </sup> Curran J and Yarus M, 1989, “Rates of aminoacyl-tRNA selection at 29 sense codons in vivo”, J Mol Biol, Vol 209, pp. 65-77. | <sup>30 </sup> Curran J and Yarus M, 1989, “Rates of aminoacyl-tRNA selection at 29 sense codons in vivo”, J Mol Biol, Vol 209, pp. 65-77. | ||
| + | |||
| + | <sup>31 </sup> Brackley C et al. 2011, “The Dynamics of Supply and Demand in mRNA Translation”, PLoS Computational Biology, Vol 7. | ||
| + | |||
| + | <sup>32 </sup> Keiler K et al. 2015, “Mechanisms of ribosome rescue in bacteria”, Nature Reviews Microbiology, Vol 13. | ||
Latest revision as of 13:07, 18 September 2015
TransOpt: A New Codon Harmonisation Model
TransOpt (Translation-Optimiser) represents a new and unique method for codon usage optimisation. It builds upon recent research and understanding, and addresses many issues inherent in the overly-simplified approaches currently taken throughout the synthetic biology community. To do this it utilises a simulated annealing approach to optimisation, and can simultaneously tailor multiple properties of any suitable gene sequence. Through optimising the translation of mRNAs in their heterologous expression hosts we hope to ensure efficient and effective expression of the desired genes in convenient and fast-growing host bacteria.
Motivation
Throughout science and industry it is frequently desirous that proteins, such as the mono-oxygenase enzyme on which our iGEM project has focused, be produced in large quantities. This often necessitates expression in non-native hosts, so as to facilitate a more convenient and efficient expression of the protein at high yields. However, the heterologous expression of a protein often remains ineffective when the wild type (WT) sequence of a gene is used directly in the expression host. In addition to approaches such as co-expression of chaperones, a range of sequence optimisation tools have been developed to overcome this,1,2 which take advantage of redundancy in the genetic code (where multiple codons encode the same amino acid (AA)) to make synonymous changes. These are generally based upon various quantisations of translation rate3 and in some cases may account for factors which impact protein expression, including mRNA secondary structure 4, 5, 1, protein post-translational modifications 1,6, differing translation rates 1, 7, and the differing tRNA abundances between organisms. 8,9
However, these approaches to optimisation frequently fall short in reflecting current understanding and research in the field, and thus fail to provide reliable optimisation with any meaningful generality. To address this, for our iGEM project we have developed a new mathematical approach to codon optimisation, TransOpt, based on the latest experimental results and theoretical understanding.
We hypothesise that, due to evolutionary pressure, gene sequences in a native host will already be optimised to provide an efficient translation process in terms of protein folding and production. As a consequence it would make sense that ribosomal initiation rate, rather than potential errors in the ensuing translation, would be the rate-limiting step in a native protein’s production: This results in a system with less energy wasted by protein misfolding, yielding a more competent organism. In order to mimic this, sequence optimisation should therefore attempt to match translational kinetics between native and heterologous hosts, whilst utilising engineering of the ribosome binding site to determine overall production rate. It is the first of these considerations that we aim to tackle with TransOpt.

Scientific Background
It was long believed that codon redundancies in the genetic code had no evolutionary significance. However, in recent years experiments have shown that the expression of proteins can vary by more than 1000-fold, depending on choices made between synonymous codons (ones which encode the same AA) at an mRNA level.4,7, 10 To address this realization, a number of mechanisms via which synonymous codon choices may impact gene expression have been proposed and tested.11
Codon Adaptation
Translation rate has been shown to correlate with protein expression via its regulatory effect on AA chain formation speed, which determines the time available for co-translational folding of a protein.12 Broadly speaking, mRNA sections in which ribosomes move slowly are useful as they allow time for folded protein domains (e.g. alpha-helices) to form 1,9 (Figure 1), which in some cases require near-stalling at critical stages of synthesis.5,13 The opposite is also true, where it is found that the production of some proteins with sections prone to misfolding may benefit from fast-translating codons to speed through areas in which the erroneous folding might, if given time, occur.14 Together, these results show that depending on the particular protein, optimizing for both regions of fast-translating and slow-translating codons (Figure 2) can be equally important.14
Whilst it is generally accepted that translation rate has a major impact on protein expression, the method by which this rate is quantised can differ substantially.3 One often employed measure is the codon bias (also referred to as codon usage frequency), which assumes the translation rate of a given codon is proportional to its abundance within the genome of an organism.15,17 However, in some cases this quantisation has shown only minor impact on a protein’s expression10,11 (though this will obviously vary between proteins), whilst in others the level of expression has been dominated by the usage frequency of a small subset of the available codons 7 . Other more recent quantisations of translation utilise both Watson-Crick (WC) and non-WC interactions, estimating rates based on the availability of appropriate tRNA species within the cell, themselves quantised by their respective Gene Copy Numbers (GCNs).8 Though these approaches have shown promising results, 9 their simplistic treatment of tRNAs (based on just their GCNs) may have weaknesses, as it fails to account for factors including differing tRNA efficiencies at a given concentration18 and the dependence on amino-acylated (charged) tRNA concentration (which relies on the amino acyl-synthase enzyme kinetics), rather than tRNA concentration as a whole. 3,7
mRNA folding
Another factor that influences protein expression is the localised folding of mRNA,3,19 (Figure 3) particularly near the ribosome binding site (RBS).1,2,5 To this end it is found that strong RBS folding reduces gene expression by inhibiting ribosome initiation,20 and in some cases this may be the deciding factor (substantially outweighing the influence of codon adaptation) in a protein’s expression level.10 mRNA folding further downstream has also demonstrated various effects1 however its relationship with gene expression is yet somewhat unclear.11
Whilst relatively straightforward estimation of 2D mRNA structure is possible it remains something of an uncertain art.21 Therefore, optimization techniques may be better served by rougher quantisations of an mRNA’s folding. One such quantisation is GC concentration 22 which has been shown to correlate with mRNA secondary structure 23 in that when it is high, strong secondary structure is more likely to form. 11
Other factors
A range of other codon-based factors bearing influence on gene expression have been proposed. 3,11
Ramps (slow-translating codons near the RBS), which serve to steadily accelerate a ribosome’s translation speed in the first ~50 codons after initiation, have been shown to be particularly pronounced in highly-expressed genes (at least, in yeast).24
Throughout mRNA, once used for the first time codons are found to occur more frequently in the subsequent nearby sequence, with this heightened usage diminishing slowly with distance. 25 It has been demonstrated that this results in a heightened translation rate (and decreased translation rate when nearby codon selection is anti-correlated), which suggests that tRNA diffusion away from the ribosome may be slower than translation: Because a given tRNA is nearby, it is expedient to utilize it for encoding of subsequent codons, as its recharge time is smaller than the wait required for a different tRNA’s arrival. 25
Adjacent pairs of amino acids, which due to codon redundancy can be encoded in as many as 36 different ways, show departure from statistical randomness. Instead, a “codon pair bias” is observed in which certain pairs occur with unexpected frequency, with under-represented pairs resulting in a substantially decreased rate of translation (and vice-versa for over-represented pairs) when other controls (such as codon bias and folding free energy) are kept equal. 26
Existing Approaches to Optimisation
As experiments have shown that translation rate (and other affects discussed above) can have a substantial impact on protein expression, it is desirous that synthetic biology develop optimisation tools to account for these deciding influences. Based on the assumption that a gene is expressed relatively efficiently in its native host, optimisation approaches therefore aim to maintain translational kinetics under the different cellular conditions of the heterologous host. Though several fairly advanced tools for codon optimisation exist 1 many authors continue to employ simple rate-maximisation or harmonisation tools when transferring a gene between hosts.
One common approach to optimisation involves selecting codons to “maximise” the translation rate, whether in terms of Codon Adaptation Index (CAI) or Codon Bias 1 in the hope of thereby maximising gene expression. However, this method frequently proves unreliable, with its failure very likely stemming from the need for slow-translation regions to allow proteins adequate time to fold. This has been experimentally demonstrated, and was confirmed by our own results (see Experimental Validation), where sequences in which fast-translating codons are selected result in substantially decreased protein activity.8
Another popular method is Codon Harmonization 15 which (in its various forms) attempts to select codons in the heterologous expression host with usage frequencies (Codon Bias) matched as near as possible with those in the native expression host. This, however, is not necessarily the best method for aligning translation rates in different mRNA regions (as will be examined in following sections), nor does it generally consider secondary effects such as mRNA structure (e.g. as approximated by GC content) or tRNA abundances.
The TransOpt approach
Though existing optimisation tools have shown some promise, they generally fail to account for a range of factors 1 6, (as discussed above), many of which have only become clear in recent years. To remedy this we have developed the TransOpt approach, which is based on the up-to-date methods for rate quantisation, and is able to simultaneously optimise any user-defined qualities of an mRNA sequence. The central optimisation methodology that we have developed is based upon a simulated annealing approach, the likes of which find widespread application in physics, mathematics, and engineering.
Quantising Codon Translation Rate
To quantise the translation rate of individual codons we utilise the methodology of Spencer et al8 which has shown promise in recent experiments regarding the impact of mRNA translation on protein folding.9This assumes that the availability of tRNA molecules within the cellular environment is the limiting factor in translation, such that when there are few tRNA species able to bond with a certain codon, it will translate slowly. In contrast to the codon bias approach to rate quantisation, which in some studies has shown negligible correlation with correct protein folding (and hence expression)10 , this methodology does not require assumptions be made about how genome-wide trends in codon usage should transfer from native to heterologous expression hosts. This, we believe, is important due to the demonstrated need for both fast- and slow-translating stretches of an mRNA 1, 12 , rendering it impossible to make assertions about individual codon rates based on their genome-wide usage (i.e. codons may be used frequently in a genome by virtue of their slowness in translation!). Instead, the methodology of Spencer et al. 8 assumes that the rate at which a codon is translated is a function of the number of (total) tRNAs able to do so, which in turn is a function of the GCN of individual tRNA species within the organism’s genome. This approach is of course not without potential downsides, such as the demonstrated dependence of rate on how highly charged a tRNA species is 7 as well as the fact that native aminoacyl-tRNAs may not be equally efficient in translation.18
Beginning with an organism’s tRNA gene content, which for many organisms is available online 27 , the rate for each individual codon is calculated as a function of tRNA species available to encode it. This process, described at length in supplementary material of the paper by Spencer et al 8 , assumes that Watson-Crick (WC) interactions are allowed between a codon and its matching tRNA species, and in some cases between a codon and a similar tRNA species whose binding is permitted due to non-WC (wobble 28) interactions. The rate for WC interactions is proportional to the specific tRNA’s abundance (i.e. GCN), whilst that for the non-WC interactions is proportional to one third of their abundance. This one third “penalizing factor” has been experimentally determined as an approximate slow-down for wobble interactions. 29,30 The rate for a given codon is then equal to the sum of contributions from to these two mechanisms, though some minor exceptions to these rules exist.8 Utilising this quantisation of rate in different organisms introduces the assumption that there is a similar number of tRNA molecules in each organism, which will be a subject of future study.
Determining Translation Rate Trends
Rather than defining translation rate based on instantaneous values for single codons, the approach of Spencer et al.8 calculates longer-term trends via a moving average centred at a given location. To take an example, the translation rate at codon location n \((r_n)\) in an mRNA sequence is equal to mean of the rates \((r_i^*)\) for individual codons between location \(n-w\) and \(n+w\), where \(w\), is the averaging half-width, which we set as \(w = 15\).8 Expressed mathematically this is:
$$r_n=\left(\sum^{n+w}_{i=n-w}{r^*_i}\right)/(2w+1)$$
We believe this approach is more physically realistic as it represents the local trend in rates, which has been shown to be an important determinant of success in protein folding. 9,14 This means that, in contrast to standard codon harmonization approaches15 it may be advantageous in some circumstances to choose a codon in the heterologous expression host possessing a substantially different rate to that in the native expression host, in order to optimise the overall trend.
An example of why this can be important is demonstrated in Figure 4. Here, a standard codon harmonization algorithm (red) would select synonymous codons with rates of 0.1 and 0.3 for codons C1 and C2 respectively, as these are the closest to the native host rates. This gives a “long term” average rate across the two of only 0.2, substantially departed from the average rate in the native host, 0.45. However, if we instead select codons so as to optimise the average rate we attain a different outcome, with the optimal approach (green) actually choosing a codon with a greater difference in rate from the original host (in this case, selecting C2 rate as 0.8, rather than 0.3, to replace the initial 0.5). Making this choice results in an average rate in the heterologous host of 0.45, exactly the same as that in the native host, despite the fact that we haven’t chosen the closest matching codon at each individual point.
An additional benefit of optimising the long-term average rate is that it will not introduce factors (discussed previously) such as codon pair bias 26 or correlation between codons utilised nearby to one another.25 These may be impacted by standard harmonization methods, where the regimented mapping between codons frequently assigns multiple native host codons to a single heterologous host codon, thereby increasing that codon’s usage substantially beyond what might be desirable.
Approximating Folding
Localised folding of mRNA has been shown to have an effect on protein expression3, 19 and in TransOpt this is approximated via measure of GC concentration, itself having been shown to correlate with mRNA secondary structure 23 We therefore include this in our optimisation algorithm, as an attempt to make mRNA folding patterns (which may influence translation kinetics) in the heterologous host similar to those in the native host.
To quantise the GC concentration we propose a metric called the GC Richness, defined in a similar way to the long-term translation rate trends discussed above, via:
$${GC}_n=\left(\sum^{n+w}_{i=n-w}{{GC}^*_i}\right)/(2w+1)$$
Here \(GC_n\) is the GC Richness (a number between \(0\) and \(1\)) at location n, and \(GC_i^*\) is the GC content of the codon at location i, defined as the proportion of that codon made up of G or C nucleotides (so, GCA would have GC content of \(2/3\), whilst TAA would have GC content of \(0\)). We take the same averaging half-width (\(w =1 5\)) as previously used for determination of the rate.
Optimisation Technique
With the defined metrics for translation rate (from here this will just be referred to as rate) and GC Richness in hand, we are ready to begin optimising the codon choices in a sequence for heterologous expression. Figure 5 presents an example of these metrics for a ~585 codon length (1755 bp length) piece of mRNA in its native expression host. This therefore forms the target profile of our optimisation algorithm: We will attempt to adjust synonymous codon usage in a new sequence such that in the heterologous expression host both the rate and GC Richness replicate those in the native expression host as near as possible.
As changes to an individual codon will alter both the rate and GC Richness at all points within width \(w\), we are unable to devise any closed-form analytical optimisation method. Instead, we utilise a technique based upon simulated annealing, a commonly used probabilistic approach to global optimisation, which relies on repeated random adjustments to slowly seek out new and better matching sequences.
Put simply, our proposed optimisation method involves defining an energy measure which is a function of both rate and GC richness, making random synonymous changes to codons, and then accepting these changes if they improve the alignment with the target profile, whilst also accepting them with some probability if they decrease alignment with the target profile. This last step, accepting inferior sequences, is included to allow the algorithm to escape local minima in the energy function, though it by no means guarantees a global minimum is ever reached.
The outer algorithm (Algorithm 1) for this approach is concerned with taking the initial codon sequence \((s_in)\), which is used to define target profiles for rate (\(r_t\)) and GC Richness (\(GC_t\)) based on the initial sequence in the natural expression host. This sequence is adjusted via Algorithm 2, and the result (\(s_0\)) is compared to the previous sequence (\(s_2\)) in the heterologous expression host whose rate (\(r_2\)) and GC Richness (\(GC_2\)) best-matched their target profiles (\(r_t\) & \(GC_t\)) from the natural expression host. If the new sequence is an improvement it keeps it (i.e. \(s_2\) is set to equal \(s_0\)), otherwise the new codon sequence is rejected in favour of the previous best sequence (i.e. \(s_2\) is not changed) and the algorithm continues.
Algorithm 1
1. Input codon sequence (\(s_{in}\)), and target rate (\(r_t\)) and GC Richness (\(GC_t\)) profiles.
2. Execute Algorithm 2 to acquire new synonymous sequence (\(s_0\)) with rate (\(r_0\)) and GC Richness (\(GC_0\)) profiles.
3. If the absolute deviation (d) of the new sequence’s rate from the target rate (i.e. \(d=\sum_n{|r_t-r_0|}\)) is smaller than all previous values of d, then accept this new sequence as the current most optimal sequence (\(s_2\)), and record its rate (\(r_2\)) and GC Richness (\(GC_2\)) profiles.
4. Repeat from step 2 using the current most optimal sequence (\(s_2\)).
Within Algorithm 1 is Algorithm 2, which runs on an internal repeat loop. Algorithm 2 is responsible for making tactical synonymous changes to the codon sequence.
Algorithm 2
1. Input codon sequence (\(s_{in}\)), target rate (\(r_t\)) and GC Richness (\(GC_t\)) profiles, and averaging width (\(k\), where \(k = 2*w + 1\) with w the averaging half-width as defined previously). Initialise offset \(i\) as \(i = 1\).
2. Randomly synonymously alter codons at locations \(i+nk\) where \(n=0,1,2...\). (so \(i,i+k,i+2k,...\)) in the initial sequence \((s_0)\) to form an altered sequence \((s_1)\).
3. Calculate new rate \((r_1)\) and GC Richness profiles \((GC_1)\) for altered codon sequence \((s_1)\).
4. Calculate the change in “energy” between the altered and original sequence at each location \(i+nk\), given by: $$\triangle E_{i+nk}=\sum^{i+nk+w}_{j=i+nk-w}{\alpha \times \left(|r_{0_j}-r_{t_j}|-|r_{1_j}-r_{t_j}|\right)+\beta \times \left(|GC_{0_j}-GC_{t_j}|-|GC_{1_j}-GC_{t_j}|\right)}+f$$
Where \(\alpha ,\ \beta\) are weighting factors for different terms in the optimisation function, in our case set to \( \alpha =1,\ \beta =0.3\), and \(f\) is an arbitrary function, in our case set as \(f = 0\).
5. At each location \(i+nk\) in \(s_0\) accept the codon change in \(s_1\) if \(\triangle E_{i+nk} \geq 0\) or if \(\triangle E_{i+nk}<0\) accept the change with probability: $$p\left(\triangle E_{i+nk}\right)=1-exp(\epsilon \times \triangle E_{i+nk})$$ where \(ϵ\) is a scaling factor, in our case set so \(ϵ=1/k\). Otherwise, reject the changed codon (let the codon at location \(i+nk\) in \(s_0\) remain the same).
6. If \(i<k\) increment \(i\) by 1 \((i=i+1)\) and repeat from step 2 using the updated sequence \(s_0\). Otherwise, end algorithm and output sequence \(s_0\) and its corresponding rate (\(r_0\)) and GC Richness (\(GC_0\)) profiles.
Conceptually, Algorithm 2 makes changes to the sequence at discrete points separated such that a change at any one will not affect the rate or GC Richness (which are averaged within a width \(w\) of a given point) at another. Then an energy value is calculated, which is used to drive the heterologous rate and GC Richness profiles towards their target profiles. Mathematically, this is equivalent to minimising the area shaded blue in the Figure 6 A about each point.
This area is calculated in the energy equation as the sum of absolute deviations between the various profiles, and is compared between the sequence with randomly altered codons (\(s_1\)) and the initial sequence (\(s_0\)). A positive energy therefore corresponds to a segment of \(r_1\) in which the shaded area is smaller in magnitude than the corresponding area about the same point in \(r_0\) (the unaltered sequence). Hence, by accepting changes that create a positive energy we are selecting sequences which reduce this absolute deviation between current and target profiles, thereby driving the heterologous profiles towards the target profiles.
This is repeated as we increment the offset (\(i\)) in step 6 and move through the sequence of codons, which is demonstrated for the first iteration in Figure 6 B.
Here we see the locations at which codons are being randomly altered (red lines) move one step to the right, and then Algorithm 2 repeats with an updated sequence (\(s_0^*\)), which was generated in the previous iteration by including some changes in \(s_1\) into \(s_0\).
Overview Flowchart
Combining Algorithms 1 and 2 we can create a summarised flow chart (Figure 7) for our optimisation procedure. It has not escaped our notice that Algorithm 1 repeats indefinitely. By design the optimiser will continually generate and display better-optimised sequences, asking the user every 30 iterations whether they would like to terminate the process.
Generalised Capabilities
Running the TransOpt algorithm with an arbitrary mRNA (in this case, luciferase 8 ), as done in Figure 8, can be illustrative for demonstration of its capabilities. A number of observations are apparent:
- The rate and GC Richness profiles for the TransOpt optimised sequence (yellow) match very closely with the target values (blue). This shows the algorithm is performing as desired: It is forcing both the rate profile and GC Richness profile to converge to their target profiles.
- The rate profile for the sequence with standard harmonization (red) does not accurately match the target profile, and in many areas is worse than the wild type sequence expressed in the heterologous host without any optimisation whatsoever.
- The GC content profile for the sequence with standard harmonization departs significantly from the target profile. The target profile would be achieved by expression in the heterologous host without adjustment, as the GC Richness is a function of the codon sequence, not organism.
- The output from TransOpt typically converges to a reasonable result within ~5 iterations, doing so in approximately 10 seconds on a standard laptop computer. However, as the user is required to stop optimisation once they deem an acceptable solution has been reached, the algorithm can run indefinitely to gradually improve its results.
Experimental Validation
Methodology
In order to better understand how the algorithm performed in achieving its mandate, it was experimentally validated via measuring the fluorescence of variants of Bacillus subtilis fluorescent protein (BsFP), different at the transcriptome but not proteomics level. This was taken from its natural host B. subtilis, and expressed in E. coli, which meant utilising the tRNA GCNs for these two organisms for rate quantisation.27 As in many other studies, the fluorescence of protein variants is assumed to be proportional to the level of protein expression and hence successful folding, which is what the algorithm aims to optimise. For more detailed explanation of the experimental design please refer to the results page.
To assess the performance of a TransOpt variant of this protein we produced four synonymous strains by different methods:
- BsFP-WT: native sequence from the native host B.subtilis
- BsFP-fast: all codons replaced with their synonyms possessing the highest rate, quantised using the approach of Spencer et al8
- BsFP-standard: harmonised BsFbFP generated via standard harmonisation (as proposed by Angov et al15 ) using the rate quantisation approach of Spencer et al8 discussed above.
- BsFP-TransOpt: optimised sequence generated using the TransOpt algorithm

In Figure 9 the translation rate and GC Richness profiles for these four sequences are presented, and a number of features are evident:
- The rate profile of the WT sequence when expressed unaltered in e coli is not actually that far from the target profile (WT in Bacillus subtilis). This implies that the difference between the two organisms, in terms of our quantisation for translation rate, is not particularly great.
- The Fast-optimised sequence has, as intended, substantially increased the translation rate at all locations in the sequence. In this case this has also resulted in an increased GC Richness throughout the sequence.
- The Standard harmonised sequence matches fairly well with the wild-type Bacillus subtilis target sequence in both profiles. Its rate profile matches more closely with the target sequence than the un-altered WT sequence when expressed in e coli, however it is outperformed by the TransOpt sequence.
- The TransOpt sequence matches closely with the target rate profile, outperforming both the standard-optimised and WT sequence, and also matches well with the target GC Richness profile.
Results
Our experimental results, presented with more technical detail on the Project Results page, are summarized in Figure 10.
Standard Codon Harmonisation
The sequence derived using standard codon harmonisation methods showed the strongest fluorescence, improving on the wild type sequence by a factor of more than 3. This result supported by previous experimental studies of fluorescence proteins, where it has been demonstrated that this commonly used harmonisation algorithm can substantially improve performance (for example, in the work of Angov et al 15). To perform this harmonisation we used the translation rate quantisation methods of Spencer et al8 , and the strong expression here therefore provides evidence for the effectiveness of these methods. In Figure 11 it is apparent that the relative utilisation of different codons in this variant is closest to that in the wild type. In Figure 9 we see that, despite the weaknesses we believe exist in this optimisation method, in this simple protein it has provided fairly well-matching translation rate and GC Richness profiles.
Fast Codon Utilisation
The fast optimised sequence presented a substantially decreased fluorescence when compared to the wild type sequence. This is also as anticipated, and agrees with previous results which have shown the necessity for both slow- and fast-translating regions in mRNA.14 In fact, our result aligns very closely with the work of Spencer et al,8 who similarly developed a synonymous sequence using the fastest-translating codons, and found its activity dropped to approximately half of its wild type performance.
A number of possible causes for this exists. For example, when using only fast-translating codons there may be insufficient time for co-translational folding of the protein, hampering its activity. However, another potential cause in this case (as examined qualitatively by our visualisation tools) is tRNA starvation: If all codons that encode a single amino acid are substituted by for a single codon species, that species' utilisation throughout the sequence will be substantially increased, potentially resulting in starvation of its corresponding tRNA species during translation.31 This is apparent in Figure 11, where it can be seen that the fast variant utilises a far smaller subset of the available codons, but uses each with a higher frequency. In the cell this might therefore be exhibited in starvation of these particular species, which is simulated by our visualisation tools. Not only can this slow translation, but may in extreme cases result in stalling, which can activate ribosomal terminating mechanisms causing premature termination of translation.32 Either of these mechanisms can cause an decrease in the cell's overall protein production, which would correspond to a decrease in fluorescence as our experimental results have demonstrated.
TransOpt
The TransOpt optimised sequence showed the weakest fluorescence, performing only marginally better than, and within experimental error of, the control measurement. This is not the result we had hoped for or expected, but as with all good science, sometimes important lessons can come from negative outcomes.
A number of potential causes for this failure exist, central among which might be our choice of test protein, BsFP. Though this comes from B. subtilis, as we have demonstrated even in its wild type form it can function effectively in e. coli, potentially due to its relative simplicity and short length. A major motivation behind TransOpt is the optimisation of sequences to account for both sections of fast- and slow-translating proteins, as well as mRNA folding. So, without a sufficiency complex protein that requires these controlling factors during translation, perhaps it is unsurprising that TransOpt would not outperform standard codon harmonisation methods. This is illustrated by Figure 9, where we see that both the WT and standard-optimised sequences do not differ very substantially from their target profiles, and hence we would expect them to perform relatively well. However, this does not explain why the fluorescence measured by TransOpt is so substantially decreased that it fails to (within error bars) outperform control measurements with non-fluorescent samples. One possible explanation may be that the TransOpt method has introduced a reliance on codon species that the heterologous expression host is unable to support.
We can identify some such species if we compare codon usage between the different protein variants. This reveals four codons (GCC, CTA, TCG and ACT, marked by red circles in Figure 11), which are not used in either the WT, Fast or Standard variants, but are used in the TransOpt variant. Therefore, if our e. coli strain was unable to effectively translate any one of these codons, for whatever reason, this could cause a failure in protein production. A potential target for investigation would be the last of these codons, ACT, for which the corresponding tRNA (AGT) in e. coli has copy number zero, and it thus must rely on the wobble interaction with other species for translation. If we examine the overall codon usage for the TransOpt sequence (Figure 11) we see that it differs more substantially from the wild type than the standard harmonisation variant, though less so than the fast variant. We can also see numerous locations where the fast and standard optimised sequence utilise codons not found in the wild type sequence, however, this did not appear to prevent their expression.
Possible Extensions
As exhibited by our experimental results, there appear to potentially be a number of shortcomings in our newly designed TransOpt approach. However, we believe that due to its scientific underpinning this method should not be discarded, and may be improved substantially by future work.
Though we believe that the failure of TransOpt to yield reliable function of BsFP may be a result of this protein's simplicity, short length, or potential errors in our fluorescence measurement experiments, the analysis of these results does highlight the possibility for tRNA starvation within the cell to play a role in protein expression. The mechanisms by which this occurs are demonstrated by our visualisation tools. Furthermore, for this particular protein the unchanged WT and standard-optimised sequences match quite well with the target profiles in Bacillus subtilis (Figure 9), leaving the TransOpt algorithm little room to make any substantial improvement.
A major improvement for TransOpt would therefore be inclusion of measures to account for this starvation: This would necessitate an additional parameter for optimisation; an expression to account for the overall usage frequency of each codon (as a total for the entire mRNA sequence, as plotted in Figure 11). Matching the ratios of these usages (as best as possible) to the tRNA GCNs would help to minimise the chance of tRNA starvation, as we would be utilising tRNAs at a rate as close as possible to the rate at which they are being produced. The astute reader will notice that in this we are proposing something similar to past efforts in codon optimisation: The matching of the codon usage between organisms. In the past this was done because codon usage was believed to correlate with rate of translation in some way. However, our motivation is substantially different: We hypothesize that codon usage instead balances with the differing rates of production between tRNA species, and in doing so regulates (and perhaps prevents) tRNA starvation.
Other possible extensions of TransOpt include improvement of parameter estimations, and addition of other metrics during optimisation:
At present the TransOpt algorithm treats the rate and GC Richness profiles, which it aims to optimise, entirely independently of another. This leaves open the possibility for added optimisation terms in the energy equation, as represented by the arbitrary function \(f\) inserted at its end. Therefore, if in the future alternate measures of an mRNA’s “fitness” are derived, which are in some way a function of its codon sequence, the algorithm can be easily extended to optimise the profile of these measures: All that needs happen is their value be calculated and fed through the algorithm, much in the same way that the GC Richness value is at present.
The weighting factors \((\alpha \ \&\ \beta )\) in the energy equation are used to determine the relative weighting of the rate and GC Richness optimisation terms. Such semi-arbitrary factors are ubiquitous in a range of optimisation techniques, as a means to scale the importance of different quantities to be optimised, and are generally (as they have been here) experimentally determined. Via greater experimentation with our algorithm it may be possible to choose better factors to replace these, which would certainly be necessary should we include any additional optimisation terms. This might serve to decrease convergence time of the algorithm, however, we found similar resultant sequences were formed when these parameters were changed, so perhaps this is unlikely to improve the algorithm's performance from a biological perspective.
Though we have used the quantisation of codon translation rate as proposed by Spencer et al 8 this choice makes no impact on the TransOpt algorithm’s mechanism. The rate quantisation is only responsible for providing data for different codon rates, which being essentially arbitrary to the algorithm’s function can be changed to anything desired. Therefore, it is possible to easily alter the TransOpt algorithm to utilise any quantisation of rate (for example, the Codon Usage Frequency / Codon Bias). That said, due to the strong fluorescence of our standard harmonised BsFP variant (which utilised the Spencer et al rate quantisation), it appears that the Spencer et al rate quantisation is functioning (at least to some extent) as intended.
Further study could also advise the choice of averaging width (\(w\)), which impacts the GC Richness and rate profiles. At present this is set to the value recommended by Spencer et al8 and is meant to reflect a characteristic length over which the translation process’s kinetics are impacted by the mRNA sequence. Decreasing this value would allow finer-resolution matching of a sequence to the target rate and GC Richness profiles, but would come at the expense of this match’s accuracy. If we set \(w=1\), then TransOpt would essentially return the same sequence as standard harmonisation tools: In each individual location it will return for the best matching rate. To properly assess this parameter’s impact greater experimental data would be necessary, which could involve further study on the interdependency of translation rate and protein folding.9
Conclusion
We have developed a new approach to codon optimisation, TransOpt, which aims to match several properties of a sequence in its heterologous expression host to that in the expression host. The mathematical approach to optimisation, simulated annealing, works exceptionally well, and demonstrates a capability to match these properties very accurately between sequences. However, experimental results demonstrated that in our test case a TransOpt optimized sequence did not function as expected: It resulted in negligible expression of a fluorescent protein. We identified a number of possible causes, such as this protein's simplicity and the fact that the TransOpt sequence utilised a number of codons not found in any of the other, more successful, variants. These variants included a sequence optimised by standard harmonisation methods, which showed a substantial increase in fluorescence over the wild type sequence, and a sequence in which only fast-translating codons were selected, which showed a substantial decrease in fluorescence compared to the wild type. These outcomes confirm previously published results regarding codon usage, and demonstrate that our rate quantisation metrics are functioning (to some degree) as intended. Finally, future improvements to the TransOpt algorithm are identified, and we suggest topics for future study which could more rigorously test certain features of TransOpt.
References
1 Angov E, 2011, “Codon usage: Nature’s roadmap to expression and folding of proteins”, Biotechnol J., Vol 6, pp 650-659.
2 Woo Seo S et al., 2013, “Synthetic biology: Tools to design microbes for the production of chemicals and fuels”, Biotechnology Advances, Vol 31, pp. 811-817.
3 Gingold H and Pilpel Y, 2011, “Determinants of translation efficiency and accuracy”, Molecular Systems Biology, Vol 7.
4 Kudla G et al., 2011, “Coding sequence determinants of gene expression in Escherichia coli”, Science, Vol 324, pp. 255-258.
5 Gloge F et al., 2014, “Co-translational mechanisms of protein maturation”, Current Opinion in Structural Biology, Vol 24, pp. 24-33.
6 Arpino J et al., 2013, “Tuning the dials of Synthetic Biology”, Microbiology, Vol 159, pp. 1236-1253.
7 Welch M et al., 2009, “Design Parameters to Control Synthetic Gene Expression in Escherichia coli”, PLoS ONE, Vol 4.
8 Spencer P et al., 2012, “Silent Substitutions Predictably Alter Translation Elongation Rates and Protein Folding Efficiencies”, Vol 422, pp. 328-335.
9 Kim S et al, 2015, “Translational tuning optimizes nascent protein folding in cells”, Science, Vol 348, pp. 444-448.
10 Gustafsson C, et al., 2004, “Codon bias and heterologous protein expression”, Trends Biotechnol, Vol 22, pp. 345-353.
11 Plotkin J and Kudla G, 2011, “Synonymous but not the same: the causes and consequences of codon bias”, Nature, Vol 12, pp. 32-42.
12 Pechmann S and Frydman J, 2013, “Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding”, Nature Structural & Molecular Biology, Vol 20, pp. 237-244.
13 Puglisi D, 2015, “The delicate dance of translation and folding”, Vol 348, pp. 399-400.
14 O’Brien E et al., 2014, “Kinetic modelling indicates that fast-translating codons can coordinate cotranslational protein folding by avoiding misfolded intermediates”, Nature Communications, Vol 5.
15 Angov E et al., 2008, “Heterologous Protein Expression Is Enhanced by Harmonizing the Codon Usage Frequencies of the Target Gene with those of the Expression Host”, PLoS One, Vol 3.
17 Huang T et al. 2011, “Analysis and Prediction of Translation Rate Based on Sequence and Functional Features of the mRNA”, PLoS One, Vol 6.
18 Forster A, 2012, “Synthetic biology challenges long-held hypotheses in translation, codon bias and transcription”, Biotechnol J., Vol 7, pp. 835-845.
19 Salis H et al., 2009, “Automated design of synthetic ribosome binding sites to control protein expression”, Vol 27, pp. 946-952.
20 Kubo, M and Imanaka T, 1989, “mRNA secondary structure in an open reading frame reduces translation efficiency in Bacillus subtilis”, J. Bacteriol, Vol 171, pp. 4080–4082.
21 McCaskill J, 1990, “The Equilibrium Partition Function and Base Pair Binding Probabilities for RNA Secondary Structure”, Biopolymers, Vol 29, pp. 1105-1119.
22 Kudla G, 2006, “High Guanine and Cytosine Content Increases mRNA Levels in Mammalian Cells”, PLoS Biology, Vol 4, pp. 933-942.
23 Gu W, Zhou T and Wilke C, 2010, “A universal trend of reduced mRNA stability near the translation-initiation site in prokaryotes and eukaryotes”, PLoS Comput. Biol, Vol 6.
24 Tuller T et al., 2010, “An Evolutionarily Conserved Mechanism for Controlling the Efficiency of Protein Translation”, Cell, Vol 141, pp. 344-354.
25 Cannarozzi G et al., 2010, “A Role for Codon Order in Translational Dynamics”, Cell, Vol 141, pp. 344-367.
26 Coleman R et al., 2008, “Virus Attenuation by Genome-Scale Changes in Codon Pair Bias”, Science, Vol 320, pp. 1784-1787.
27 Chan, P and Lowe T, 2009 “GtRNAdb: A database of transfer RNA genes detected in genomic sequence.” Nucl. Acids Res, Vol 37, [http://gtrnadb.ucsc.edu/ Online Database].
28 Campbell N, 2011, “Biology (Ninth ed.)”, Glenview, ISBN 0321558235, pp. 339–342.
29 Pedersen S, 1984, “Escherichia coli ribosomes translate in vivo with variable rate”, Embo J, Vol 3.
30 Curran J and Yarus M, 1989, “Rates of aminoacyl-tRNA selection at 29 sense codons in vivo”, J Mol Biol, Vol 209, pp. 65-77.
31 Brackley C et al. 2011, “The Dynamics of Supply and Demand in mRNA Translation”, PLoS Computational Biology, Vol 7.
32 Keiler K et al. 2015, “Mechanisms of ribosome rescue in bacteria”, Nature Reviews Microbiology, Vol 13.