Difference between revisions of "Team:Pretoria UP"
| (110 intermediate revisions by 2 users not shown) | |||
| Line 20: | Line 20: | ||
#animation | #animation | ||
{ | { | ||
| + | padding-top:140px; | ||
| + | padding-left:5%; | ||
| + | } | ||
| − | top: | + | #bannerSpace |
| + | { | ||
| + | top: 4.7em; | ||
| + | width:100%; | ||
} | } | ||
| + | |||
| Line 31: | Line 38: | ||
height: 0px; | height: 0px; | ||
| − | opacity: 0. | + | opacity: 0.8; |
background: black; | background: black; | ||
} | } | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</style> | </style> | ||
| Line 50: | Line 50: | ||
window.onload = function() { | window.onload = function() { | ||
performMenuHiding(); | performMenuHiding(); | ||
| − | $('#animation').css({marginTop: '-= | + | $('#animation').css({marginTop: '+=150px'}); |
| + | |||
| + | //$('#bannerHolder').css({marginTop: '-=130px'}); | ||
| + | |||
| + | |||
}; | }; | ||
| Line 58: | Line 62: | ||
<body> | <body> | ||
<div class="wtf_div"> | <div class="wtf_div"> | ||
| + | <div align="center" id ="bannerHolder" > | ||
| − | + | <img id="Banner" alt="Banner By Natascha Muller" src="/wiki/images/0/03/Banner_Pretoria_UP.png" style="width:30%; padding-top:7.5%;"/> | |
| − | <img id="Banner" alt="Banner By Natascha Muller" src="/wiki/images/ | + | |
| + | |||
| + | <!--<h4 id="description">SYNCHRONIZED, CONDITIONAL, GENETIC CHEMOTAXIS PROGRAMMING</h4>--> | ||
| + | <h2 id="description"> Synchronized, Conditional, Genetic Chemotaxis Programming </h2> | ||
| − | |||
</div> | </div> | ||
| − | <div align="center" style="width:90%;height: | + | <div align="center" style="width:90%;height:840px;"> |
| − | + | ||
| − | <img id="animation" alt="Animation by Schae Ind" src="/wiki/images/1/15/Up_animation.gif"/> | + | <img id="animation" alt="Animation by Schae Ind" src="/wiki/images/1/15/Up_animation.gif" /> |
| Line 77: | Line 84: | ||
<p> | <p> | ||
| − | + | The Pretoria UP iGEM team of 2015 have proposed a design for intelligent motile bacteria based on a combination of DNA modules that confer different functions. The synchronous behaviour of the bacteria will be achieved through a quorum sensing module allowing the population to behave as a swarm instead of independent units. A conditional response to an environmental signal is conferred through a post transcriptional control mechanism known as riboswitching, thus the bacterial swarm may recognise and respond to a chemical attractant. Since the system makes use of several modular components, an AND gate is required to process the various signals which in turn would trigger a genetic switch in the bacterial chemotaxis. The irreversible change is programmed by a DNA recombination switch which inverts the directionality of a promoter through the Cre-loxP pathway. We invite you to explore the various aspects of the project on this site. | |
</p> | </p> | ||
Latest revision as of 22:00, 18 September 2015
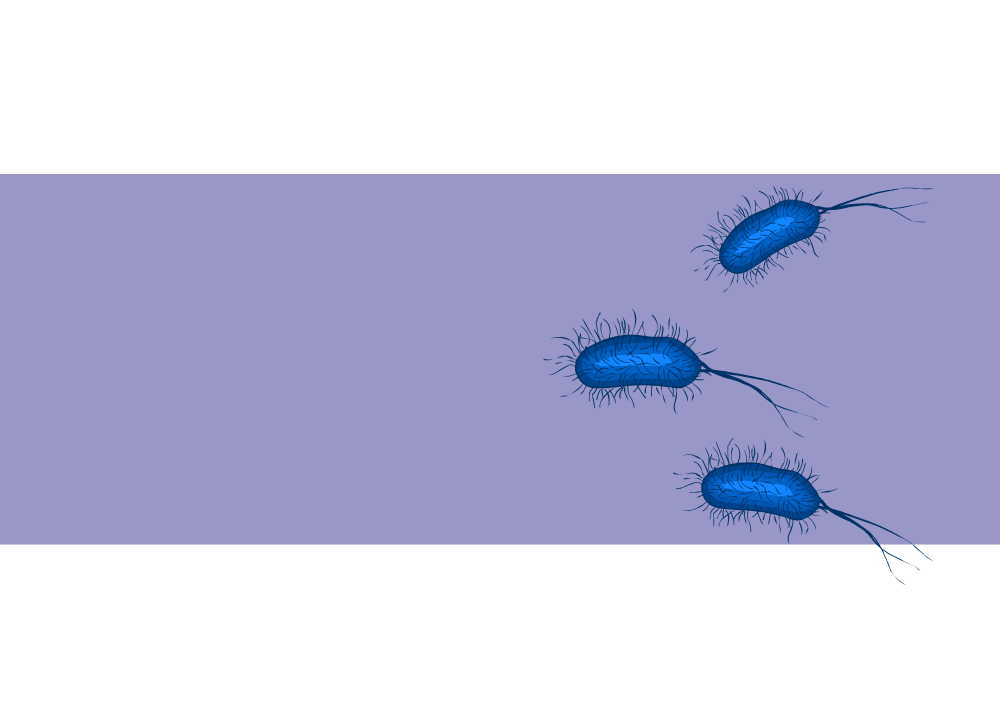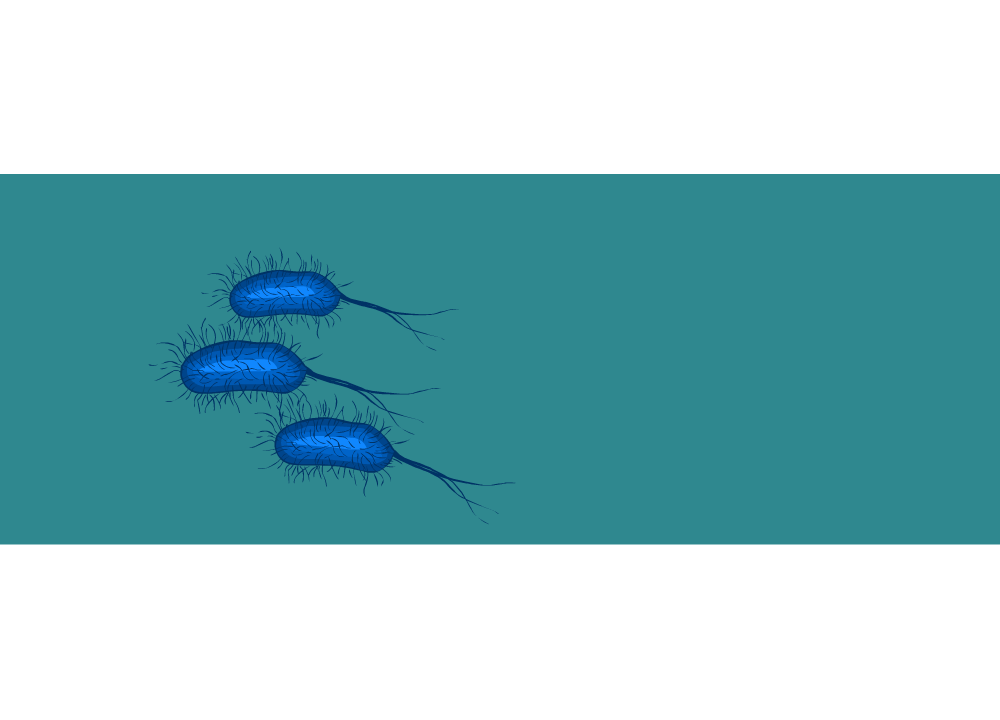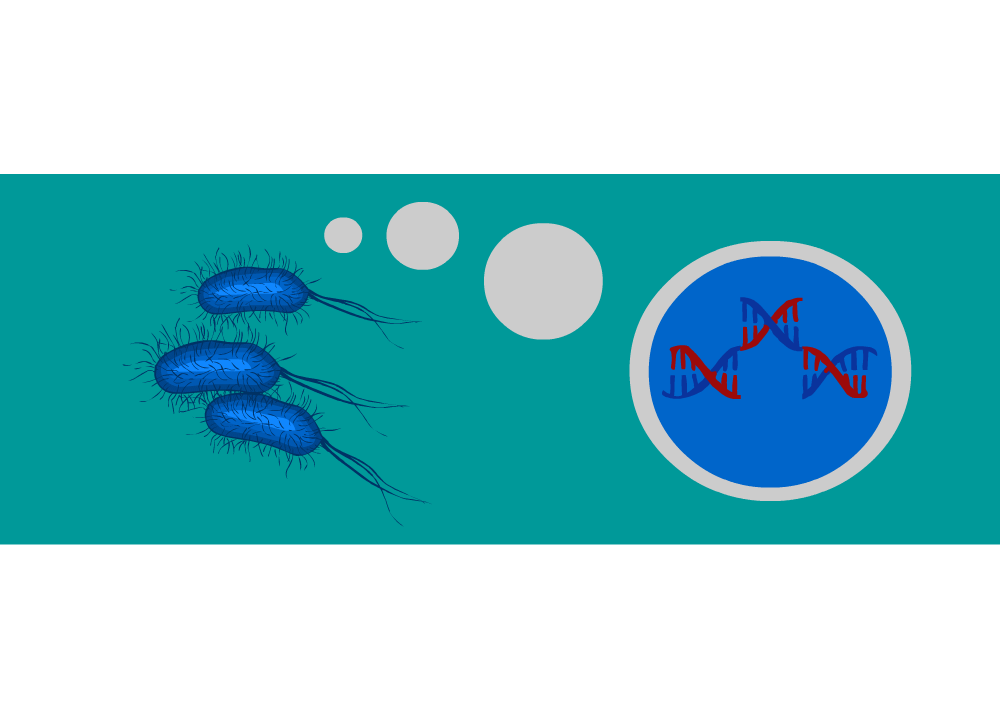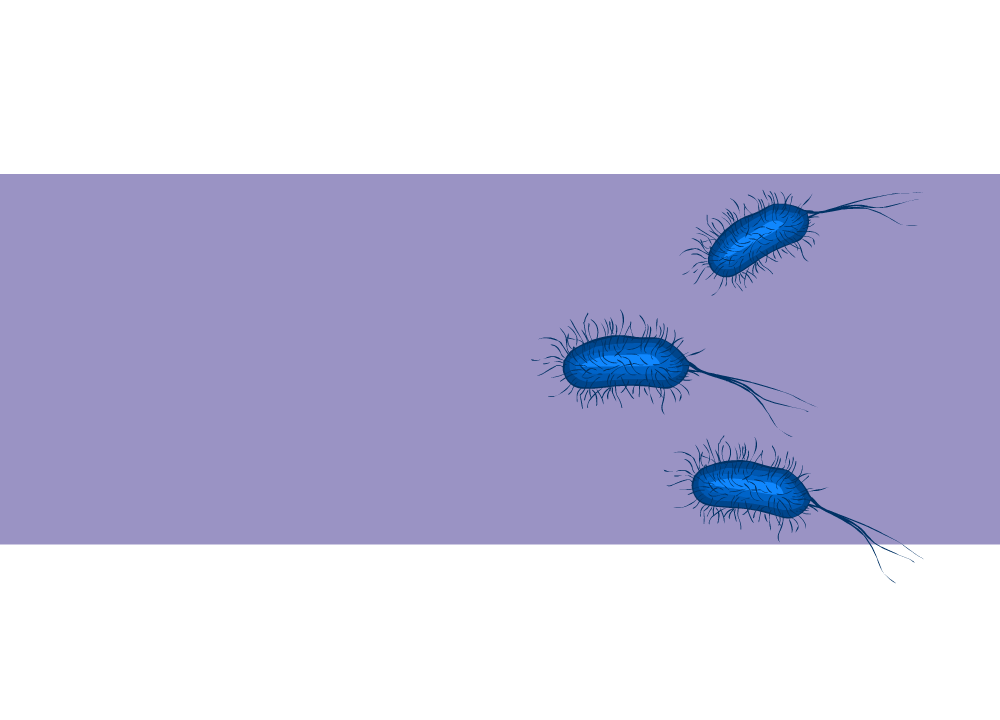
Project Description
The Pretoria UP iGEM team of 2015 have proposed a design for intelligent motile bacteria based on a combination of DNA modules that confer different functions. The synchronous behaviour of the bacteria will be achieved through a quorum sensing module allowing the population to behave as a swarm instead of independent units. A conditional response to an environmental signal is conferred through a post transcriptional control mechanism known as riboswitching, thus the bacterial swarm may recognise and respond to a chemical attractant. Since the system makes use of several modular components, an AND gate is required to process the various signals which in turn would trigger a genetic switch in the bacterial chemotaxis. The irreversible change is programmed by a DNA recombination switch which inverts the directionality of a promoter through the Cre-loxP pathway. We invite you to explore the various aspects of the project on this site.



