Difference between revisions of "Team:Stockholm/Collaborations"
Utsakarmakar (Talk | contribs) |
|||
| (101 intermediate revisions by 7 users not shown) | |||
| Line 3: | Line 3: | ||
<html> | <html> | ||
<head> | <head> | ||
| − | <link rel="stylesheet" href="https://2015.igem.org/Team: | + | <link rel="stylesheet" href="https://2015.igem.org/Team:Stockholm/bootstrap?action=raw&ctype=text/css" type="text/css"> |
| − | + | <link rel="stylesheet" href="https://2015.igem.org/Team:Stockholm/hugicss?action=raw&ctype=text/css" type="text/css"> | |
| − | + | <script src="https://2015.igem.org/Team:Stockholm/bootstrapjs?action=raw&ctype=text/javascript" type="text/javascript" ></script> | |
| + | <link rel="stylesheet" href="https://2015.igem.org/Team:Stockholm/bootstrap-lightbox.min.css?action=raw&ctype=text/css" type="text/css"> | ||
| + | <script src="https://2015.igem.org/Team:Stockholm/bootstrap-lightbox.min.js?action=raw&ctype=text/javascript" type="text/javascript" ></script> | ||
</head> | </head> | ||
<body> | <body> | ||
<div class="container-fluid content-container"> | <div class="container-fluid content-container"> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2015/b/be/Igem_collab_map_color.png" alt="Collaboration map" class="img-responsive"/> | ||
<h1> Collaborations with other iGEM teams</h1> | <h1> Collaborations with other iGEM teams</h1> | ||
| − | < | + | <p>During the year we have been involved in many extensive collaborations. We have hosted an iGEM conference for the Nordic countries, had scientific collaborations with BGU Israel, ETH Zürich, University of Freiburg, written articles for the Amoy newsletter, shared our experience in cloning in TU Eindhoven's Cloning Guide, helped CGU Taiwan with modeling, worked on digital collaboration platforms with Pasteur Paris and Aalto-Helsinki and participated in a global collaboration on human practice surveys. |
| + | |||
| + | <!-- Floating nav menu --> | ||
| + | |||
| + | <div id="sticky-anchor"></div> | ||
| + | <div id="sticky"> | ||
| + | <ul class="nav nav-tabs pillnav"> | ||
| + | <li role="presentation" class="active"><a href="#"><b> Collaborations</b></a></li> | ||
| + | |||
| + | <li role="presentation" class="dropdown anchor_link"> | ||
| + | <a class="dropdown-toggle" data-toggle="dropdown" href="#" role="button" aria-haspopup="true" aria-expanded="false"> | ||
| + | Nordic iGEM<span class="caret"></span> | ||
| + | </a> | ||
| + | <ul class="dropdown-menu"> | ||
| + | <li role="presentation" class="clickable anchor_link"><a href="#nicsummary">NiC summary</a></li> | ||
| + | <li role="presentation" class="clickable anchor_link"><a href="https://2015.igem.org/Team:Stockholm/NIC">NiC full report</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | |||
| + | <li role="presentation" class="dropdown anchor_link"> | ||
| + | <a class="dropdown-toggle" data-toggle="dropdown" href="#" role="button" aria-haspopup="true" aria-expanded="false"> | ||
| + | Scientific <span class="caret"></span> | ||
| + | </a> | ||
| + | <ul class="dropdown-menu"> | ||
| + | <li role="presentation" class="clickable anchor_link"><a href="#collab_bgu">BGU Israel</a></li> | ||
| + | <li role="presentation" class="clickable anchor_link"><a href="#collab_cgu">CGU Taiwan</a></li> | ||
| + | <li role="presentation" class="clickable anchor_link"><a href="#collab_freiburg">University of Freiburg</a></li> | ||
| + | <li role="presentation" class="clickable anchor_link"><a href="#collab_zurich">ETH Zürich</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | |||
| + | <li role="presentation" class="dropdown anchor_link"> | ||
| + | <a class="dropdown-toggle" data-toggle="dropdown" href="#" role="button" aria-haspopup="true" aria-expanded="false"> | ||
| + | Human practices <span class="caret"></span> | ||
| + | </a> | ||
| + | <ul class="dropdown-menu"> | ||
| + | <li role="presentation" class="clickable anchor_link"><a href="#collab_aalto">Aalto-Helsinki and Institut Pasteur</a></li> | ||
| + | <li role="presentation" class="clickable anchor_link"><a href="#collab_amoy">Amoy</a></li> | ||
| + | <li role="presentation" class="clickable anchor_link"><a href="#collab_surveys">Global human practice surveys</a></li> | ||
| + | <li role="presentation" class="clickable anchor_link"><a href="#collab_eindhoven">TU Eindhoven</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | |||
| + | </ul> | ||
| + | </div> | ||
| + | |||
| + | <span class="anchor" id="nicsummary"></span> | ||
| + | <h1>Nordic iGEM Conference</h1> | ||
| + | |||
| + | <p>In the collaboration with Uppsala's iGEM team, we organized and hosted a meet-up for 60 students from eight Nordic iGEM teams. Teams participating in the Nordic iGEM Conference were Oslo, SDU Danmark, Aalto Helsinki, Linköping, Copenhagen, Gothenbug as well as Uppsala and us, Stockholm. We also had a pleasure to have Randy Rettberg, president of the iGEM foundation, with us during the conference.</p> | ||
| + | |||
| + | <span class="anchor" id="collab_nic"></span> | ||
| + | <img src="https://static.igem.org/mediawiki/2015/c/c3/NiC_Header.png" alt="NIC" class="img-responsive padded-image"/> | ||
| + | |||
| + | <p> Our Nordic iGEM Conference was a three-day-long event. We had a mini-Jamboree along with the project presentations and a prize – <a href="https://twitter.com/iGEMsthlm/status/624950330799837184">the famous Golden Pipette.</a> There were also workshops, fabulous dinners and a great bar-crawl in Uppsala. It was a great experience, and the Nordic iGEM Conference will be certainly repeated next year.</p> | ||
| + | |||
| + | <h4><a href="https://2015.igem.org/Team:Stockholm/NIC">Read more about the Nordic iGEM Conference and see pictures</a></h4> | ||
| − | < | + | <h1>Scientific collaborations</h1> |
| + | <span class="anchor" id="collab_bgu"></span> | ||
<h2>BGU Israel</h2> | <h2>BGU Israel</h2> | ||
| − | <p>Together with BGU Israel we have worked on a possible | + | <p>Together with BGU Israel we have worked on a possible affibody-based application of their idea for cancer therapy. Their system is delivered to mammalian cells with Adeno-associated virus (AAV) vectors and triggers expression or knockout when two cancer-related promoters are simultaneously highly active. One method of the therapy could be to specifically target cancer cells with a toxin. Since affibody molecules can be engineered to bind almost any protein with high specificity and affinity, it might be possible to direct these toxins towards cells that express affibody molecules on their surface. To try this concept, we decided that our team would design a membrane-bound affibody that BGU Israel would then attempt to express on the surface of mammalian cells using their AAV vectors.</p> |
| − | <p>Our design | + | <p>Our design utilized a system of Glycosylphosphatidylinisotol (GPI)-anchoring that is common in membrane proteins of mammalian cells. We based our design on a human protein Decay accelerating factor (DAF), encoded by the CD55 gene. A 34-amino-acid-long signal peptide at the N-terminus precedes the actual protein, which is then followed by a second 28-amino-acid-long signal peptide at the C-terminus. The N-terminus signal directs the protein to the endoplasmic reticulum (ER), and the C-terminus signal is removed in the ER and then replaced with a GPI anchor. In addition, a DAF-derived signal has been shown to lead to GPI anchoring of fusion proteins.<a href="#ref-gpianchors"><sup class="ref"></sup></a></p> |
<img src="https://static.igem.org/mediawiki/2015/c/c5/Slide09.png" alt="BGU DAF descriptionl" class="img-responsive padded-image"/> | <img src="https://static.igem.org/mediawiki/2015/c/c5/Slide09.png" alt="BGU DAF descriptionl" class="img-responsive padded-image"/> | ||
| − | <p> | + | <p>Subsequently, we have also added a short linker sequence and a FLAG-tag on the N-terminus of the affibody. On the C-terminus, we have also added a linker that ended with serine, which is the site of cleavage for the signal peptide. This gene was then synthesized as a gBlock by IDT. The construct was expressed by the BGU Israel team. While expression of the Affibody was confirmed, it did not appear to be localized to the membrane. <a href="https://2015.igem.org/Team:BGU_Israel/Collaborations">More results are available on the BGU Israel 2015 wiki</a>.</p> |
| − | + | ||
<img src="https://static.igem.org/mediawiki/2015/e/e0/Slide10.png" alt="BGU construct description" class="img-responsive padded-image"/> | <img src="https://static.igem.org/mediawiki/2015/e/e0/Slide10.png" alt="BGU construct description" class="img-responsive padded-image"/> | ||
| − | <h2> | + | <span class="anchor" id="collab_cgu"></span> |
| + | <h2>CGU Taiwan</h2> | ||
| + | <p>The CGU Taiwan team was referred to us by the Delft University of Technology team. They needed help with the modeling part of their project. They had to knock out a gene in the G protein coupled receptor (GPCR) pathway, which in normal circumstances would be active. We introduced them to Matlab and SimBiology as well and helped them model the GPCR pathway with the eliminated gene. They were also interested in projecting the boundary condition for liquid-liquid interphase diffusion, using partial differential equations. We offered to research their reactions and to try to find kinetic parameters of the processes present in the CGU’s current project. We suggested utilization of Newmann no-flux partial differential equation useful as a boundary condition for this type of diffusion, previously used also by ETH Zürich iGEM team 2012.</p> | ||
| + | <img src="https://static.igem.org/mediawiki/2015/3/3f/Patway_design.png" alt="Design of the pathway performed by Stockholm iGEM team" class="img-responsive"/> | ||
| + | <p>Design of the pathway performed by Stockholm iGEM team</p> | ||
| − | < | + | <span class="anchor" id="collab_freiburg"></span> |
| + | <h2>University of Freiburg</h2> | ||
| − | < | + | <img src="https://static.igem.org/mediawiki/2015/3/3e/Freiburg-2.jpg" alt="Direct antigen detection using immobilzed Affibody molecules on a Ni-NTA surface." class="img-responsive"/> |
| − | <p> | + | <p>In the context of developing novel technologies for diseases diagnostics, we contacted Freiburg iGEM team who are developing a DNA Microarray to screen for several different diseases at the same time. Their project is based on the coating of a nickel surface with a certain disease antigen and the binding to the antibodies of a patient’s serum. In our collaboration, we have sent the team in Freiburg His-tagged HER-2 affibody molecules, a strong molecular binder for the breast cancer antigen. This will allow to screen directly for currently present antigens in the patient as the indirect detection via serum antibodies can also give false positive results due to old infections. |
| − | + | </p> | |
| + | |||
| + | <span class="anchor" id="collab_zurich"></span> | ||
| + | <h2>ETH Zürich</h2> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2015/c/cd/Zurich-2.jpg" alt="Testing binding capacity of BAR-expressing cells to HER-2 positive cancer cells." class="img-responsive"/> | ||
| + | |||
| + | <p>The project of ETH Zürich is in the field of cancer cell detection. Apart from several fruitful Skype conversations in which both groups have been helping each other with advice and reflections, we have also established a scientific collaboration. We characterized six of Zürich's biobricks which are different lactate sensing circuits. We tested the circuit with pure lactate and looked at their GFP production in response to increasing lactate concentrations (0.125-160 mM). In return, the iGEM Group ETH Zürich offered to investigate on binding capacity of our BAR expressing TOP10 strains to HER-2 expressing cancer cells (SK-BR-3). The attached <i>E.Coli</i> were also expressing genomic GFP. Hence, using confocal fluorescence microscopy, they tried to image BAR-expressing cells at the outside of HER-2 cells.</p> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2015/1/1f/Wiki_Image.png" alt="Testing binding capacity of BAR-expressing cells to HER-2 positive cancer cells_experiment." class="img-responsive"/> | ||
| + | <div class="figure-text"><p> Testing our three BAR expressing constructs (BAR1 (BBa_K17009), BAR2 (BBa_K1766010) and BAR3 (BBa_K1766011)) for their binding capacity towards HER-2 expressing breast cancer cells. BAR expression was initiated by ITPG induction.</p></div> | ||
| + | |||
| + | <p>In their attempt to visualize BAR expressing bacteria attached at the surface of SK-BR-3 cells, they tried to co-transform our bacteria with a plasmid expressing continuously GFP. However, this unfortunately failed. Even transforming <i>E.Coli</i>, which are stably expressing GFP with our BAR-containing plasmid seem to be ineffective. The iGEM Team ETH Zurich visualized the HER-2 expressing cells together with IPTG induced or non-induced bacteria. The results obtained due to inefficient contrast of bacteria and background remained elusive (see figure above). We hope that the increased presence of IPTG-induced bacteria in comparison to non-induced bacteria might be a sign for an increased retainment of the bacteria due to the BAR expression. However, further investigations with a slightly changed expression construct would need to be done for this purpose.</p> | ||
| + | |||
| + | <h1>Human practice collaborations</h1> | ||
| + | |||
| + | <span class="anchor" id="collab_amoy"></span> | ||
| + | <h2>Amoy</h2> | ||
| + | |||
| + | <p>Last year the iGEM team from Xiamen University together with the team from Paris Bettencourt University founded the Newsletter platform. Through this, iGEM teams from all over the world could share information about their project and discuss issues concerning it and synthetic biology in general. For this year’s version of the Newsletter iGEM Amoy contacted iGEM Stockholm to include us in the 2015 issue. We contributed with five articles concerning <a href="https://static.igem.org/mediawiki/2015/a/ae/2015-Newsletter-No.1.pdf">setting-up a team</a>, <a href="https://static.igem.org/mediawiki/2015/5/56/2015-Newsletter-No.2.pdf">germline modifications</a>, <a href="https://static.igem.org/mediawiki/2015/6/61/2015-Newsletter-No.3.pdf">a description of our project</a>, an <a href="https://static.igem.org/mediawiki/2015/c/ca/2015-Newsletter-No.7.pdf">update on our progress</a> and a <a href="https://static.igem.org/mediawiki/2015/c/ca/2015-Newsletter-No.7.pdf">controversial comment on impact factors in science.</a></p> | ||
| + | <span class="anchor" id="collab_surveys"></span> | ||
<h2>Global collaboration on human practice surveys</h2> | <h2>Global collaboration on human practice surveys</h2> | ||
| Line 51: | Line 136: | ||
<li> Tianjin University (Tianjin)</li> | <li> Tianjin University (Tianjin)</li> | ||
<li> Université de Toulouse (Toulouse)</li> | <li> Université de Toulouse (Toulouse)</li> | ||
| − | <li> University College London (UCL) | + | <li> University College London (UCL) </li> |
</ul> | </ul> | ||
</p> | </p> | ||
| − | + | <p>Our role was to come up with questions for a survey and coordinate the technical work on the survey. In a related collaboration, we also hosted a survey on our servers for iGEM Nankai and provided them with technical support.</p> | |
| − | <p> | + | |
| − | < | + | <span class="anchor" id="collab_aalto"></span> |
| − | </ | + | <h2 id="collab_aalto">Aalto-Helsinki and Institut Pasteur</h2> |
| − | < | + | <p>In the Stockholm team, several members expressed their will not to use Facebook for communication. The reasons ranged from personal to the fact that Facebook lacks key functions for streamlined collaboration and possibilities to customize it. Discussions regarding other means of communication started and the idea of the Hub arose as a common place for teams to discuss and collaborate both within the team and with other teams.</p> |
| − | <p> | + | <p>Aalto-Helsinki assumed these concerns were not limited to Stockholm and Alto-Helsinki decided to find out what other teams thought. Discussions between Stockholm, Alto-Helsinki and Institut Pasteur led to the setting up of a network using an open source platform called HumHub. This platform could potentially be used by all iGEM teams to streamline collaboration and communication within iGEM.</p> |
| − | </ | + | |
| + | <h4><a href="https://2015.igem.org/Team:Stockholm/humhub">Read the full HumHub article, written by Stockholm and Alto-Helsinki, here</a></h4> | ||
| − | |||
| − | < | + | <span class="anchor" id="collab_eindhoven"></span> |
| − | + | <h2>TU Eindhoven</h2> | |
| − | < | + | <p>Cloning methods represent one of the core elements of synthetic biology and it is crucial for each iGEM team to know as much as possible about the broad varity of cloning techniques. This need has been recognized by the iGEM Eindhoven which searched for collaborators using different cloning approaches to <a href="https://2015.igem.org/Team:TU_Eindhoven/Collaborations/CloningGuide"> share their experiences in the latest cloning guide.</a> The iGEM Team Stockholm could therefore contribute with their experience in overlap-extension PCR (OE-PCR) cloning. This technique is a completely restriction enzyme free approach to integrate elongated fragments into a target vector.</p> |
| + | <h4>References</h4> | ||
| + | |||
| + | <table class="table table-condensed reference-table"> | ||
| + | |||
| + | <thead> | ||
| + | <tr> | ||
| + | <th class="col-md-1 table-head">#</th> | ||
| + | <th class="col-md-10 table-head">Reference</th> | ||
| + | </tr> | ||
| + | </thead> | ||
| + | |||
| + | <tr> | ||
| + | <td class="col-md-1 ">1</td> | ||
| + | <td class="col-md-10 "><a href="http://www.ncbi.nlm.nih.gov/pubmed/8380601"> | ||
| + | <span class="anchor-ref" id="ref-gpianchors"></span> | ||
| + | Hannan LA, Lisanti MP, Rodriguez-boulan E, Edidin M. Correctly sorted molecules of a GPI-anchored protein are clustered and immobile when they arrive at the apical surface of MDCK cells. J Cell Biol. 1993;120(2):353-8.</a></td> | ||
| + | </td> | ||
| + | </tr> | ||
| + | |||
| + | </table> | ||
</div> | </div> | ||
| + | |||
| + | <script> | ||
| + | function sticky_relocate() { | ||
| + | var window_top = $(window).scrollTop(); | ||
| + | var div_top = $('#sticky-anchor').offset().top; | ||
| + | if (window_top > div_top) { | ||
| + | $('#sticky').addClass('stick'); | ||
| + | } else { | ||
| + | $('#sticky').removeClass('stick'); | ||
| + | } | ||
| + | } | ||
| + | |||
| + | $(function () { | ||
| + | $(window).scroll(sticky_relocate); | ||
| + | sticky_relocate(); | ||
| + | }); | ||
| + | |||
| + | $(".nav li").on("click", function() { | ||
| + | $(".nav li").removeClass("active"); | ||
| + | $(this).addClass("active"); | ||
| + | }); | ||
| + | </script> | ||
</body> | </body> | ||
</html> | </html> | ||
{{:Team:Stockholm/newfooter}} | {{:Team:Stockholm/newfooter}} | ||
Latest revision as of 22:19, 18 September 2015

Collaborations with other iGEM teams
During the year we have been involved in many extensive collaborations. We have hosted an iGEM conference for the Nordic countries, had scientific collaborations with BGU Israel, ETH Zürich, University of Freiburg, written articles for the Amoy newsletter, shared our experience in cloning in TU Eindhoven's Cloning Guide, helped CGU Taiwan with modeling, worked on digital collaboration platforms with Pasteur Paris and Aalto-Helsinki and participated in a global collaboration on human practice surveys.
Nordic iGEM Conference
In the collaboration with Uppsala's iGEM team, we organized and hosted a meet-up for 60 students from eight Nordic iGEM teams. Teams participating in the Nordic iGEM Conference were Oslo, SDU Danmark, Aalto Helsinki, Linköping, Copenhagen, Gothenbug as well as Uppsala and us, Stockholm. We also had a pleasure to have Randy Rettberg, president of the iGEM foundation, with us during the conference.

Our Nordic iGEM Conference was a three-day-long event. We had a mini-Jamboree along with the project presentations and a prize – the famous Golden Pipette. There were also workshops, fabulous dinners and a great bar-crawl in Uppsala. It was a great experience, and the Nordic iGEM Conference will be certainly repeated next year.
Read more about the Nordic iGEM Conference and see pictures
Scientific collaborations
BGU Israel
Together with BGU Israel we have worked on a possible affibody-based application of their idea for cancer therapy. Their system is delivered to mammalian cells with Adeno-associated virus (AAV) vectors and triggers expression or knockout when two cancer-related promoters are simultaneously highly active. One method of the therapy could be to specifically target cancer cells with a toxin. Since affibody molecules can be engineered to bind almost any protein with high specificity and affinity, it might be possible to direct these toxins towards cells that express affibody molecules on their surface. To try this concept, we decided that our team would design a membrane-bound affibody that BGU Israel would then attempt to express on the surface of mammalian cells using their AAV vectors.
Our design utilized a system of Glycosylphosphatidylinisotol (GPI)-anchoring that is common in membrane proteins of mammalian cells. We based our design on a human protein Decay accelerating factor (DAF), encoded by the CD55 gene. A 34-amino-acid-long signal peptide at the N-terminus precedes the actual protein, which is then followed by a second 28-amino-acid-long signal peptide at the C-terminus. The N-terminus signal directs the protein to the endoplasmic reticulum (ER), and the C-terminus signal is removed in the ER and then replaced with a GPI anchor. In addition, a DAF-derived signal has been shown to lead to GPI anchoring of fusion proteins.

Subsequently, we have also added a short linker sequence and a FLAG-tag on the N-terminus of the affibody. On the C-terminus, we have also added a linker that ended with serine, which is the site of cleavage for the signal peptide. This gene was then synthesized as a gBlock by IDT. The construct was expressed by the BGU Israel team. While expression of the Affibody was confirmed, it did not appear to be localized to the membrane. More results are available on the BGU Israel 2015 wiki.

CGU Taiwan
The CGU Taiwan team was referred to us by the Delft University of Technology team. They needed help with the modeling part of their project. They had to knock out a gene in the G protein coupled receptor (GPCR) pathway, which in normal circumstances would be active. We introduced them to Matlab and SimBiology as well and helped them model the GPCR pathway with the eliminated gene. They were also interested in projecting the boundary condition for liquid-liquid interphase diffusion, using partial differential equations. We offered to research their reactions and to try to find kinetic parameters of the processes present in the CGU’s current project. We suggested utilization of Newmann no-flux partial differential equation useful as a boundary condition for this type of diffusion, previously used also by ETH Zürich iGEM team 2012.

Design of the pathway performed by Stockholm iGEM team
University of Freiburg

In the context of developing novel technologies for diseases diagnostics, we contacted Freiburg iGEM team who are developing a DNA Microarray to screen for several different diseases at the same time. Their project is based on the coating of a nickel surface with a certain disease antigen and the binding to the antibodies of a patient’s serum. In our collaboration, we have sent the team in Freiburg His-tagged HER-2 affibody molecules, a strong molecular binder for the breast cancer antigen. This will allow to screen directly for currently present antigens in the patient as the indirect detection via serum antibodies can also give false positive results due to old infections.
ETH Zürich

The project of ETH Zürich is in the field of cancer cell detection. Apart from several fruitful Skype conversations in which both groups have been helping each other with advice and reflections, we have also established a scientific collaboration. We characterized six of Zürich's biobricks which are different lactate sensing circuits. We tested the circuit with pure lactate and looked at their GFP production in response to increasing lactate concentrations (0.125-160 mM). In return, the iGEM Group ETH Zürich offered to investigate on binding capacity of our BAR expressing TOP10 strains to HER-2 expressing cancer cells (SK-BR-3). The attached E.Coli were also expressing genomic GFP. Hence, using confocal fluorescence microscopy, they tried to image BAR-expressing cells at the outside of HER-2 cells.
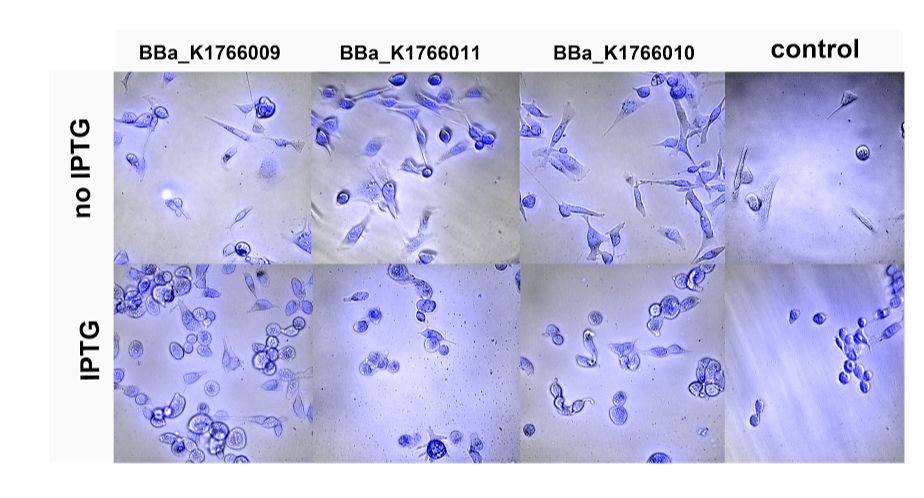
Testing our three BAR expressing constructs (BAR1 (BBa_K17009), BAR2 (BBa_K1766010) and BAR3 (BBa_K1766011)) for their binding capacity towards HER-2 expressing breast cancer cells. BAR expression was initiated by ITPG induction.
In their attempt to visualize BAR expressing bacteria attached at the surface of SK-BR-3 cells, they tried to co-transform our bacteria with a plasmid expressing continuously GFP. However, this unfortunately failed. Even transforming E.Coli, which are stably expressing GFP with our BAR-containing plasmid seem to be ineffective. The iGEM Team ETH Zurich visualized the HER-2 expressing cells together with IPTG induced or non-induced bacteria. The results obtained due to inefficient contrast of bacteria and background remained elusive (see figure above). We hope that the increased presence of IPTG-induced bacteria in comparison to non-induced bacteria might be a sign for an increased retainment of the bacteria due to the BAR expression. However, further investigations with a slightly changed expression construct would need to be done for this purpose.
Human practice collaborations
Amoy
Last year the iGEM team from Xiamen University together with the team from Paris Bettencourt University founded the Newsletter platform. Through this, iGEM teams from all over the world could share information about their project and discuss issues concerning it and synthetic biology in general. For this year’s version of the Newsletter iGEM Amoy contacted iGEM Stockholm to include us in the 2015 issue. We contributed with five articles concerning setting-up a team, germline modifications, a description of our project, an update on our progress and a controversial comment on impact factors in science.
Global collaboration on human practice surveys
CGU Taiwan involved the Stockolm team into a broader collaboration, evaluating public recognition of iGEM projects. Except for CGU Taiwan and Stockholm iGEM teams, the following teams collaborated on this:
- Technische Universität Berlin (Berlin)
- Beijing Institute of Technology (BIT)
- University of São Paulo (Brasil_USP)
- The Chinese University of Hong Kong
- University of Leicester
- Tianjin University (Tianjin)
- Université de Toulouse (Toulouse)
- University College London (UCL)
Our role was to come up with questions for a survey and coordinate the technical work on the survey. In a related collaboration, we also hosted a survey on our servers for iGEM Nankai and provided them with technical support.
Aalto-Helsinki and Institut Pasteur
In the Stockholm team, several members expressed their will not to use Facebook for communication. The reasons ranged from personal to the fact that Facebook lacks key functions for streamlined collaboration and possibilities to customize it. Discussions regarding other means of communication started and the idea of the Hub arose as a common place for teams to discuss and collaborate both within the team and with other teams.
Aalto-Helsinki assumed these concerns were not limited to Stockholm and Alto-Helsinki decided to find out what other teams thought. Discussions between Stockholm, Alto-Helsinki and Institut Pasteur led to the setting up of a network using an open source platform called HumHub. This platform could potentially be used by all iGEM teams to streamline collaboration and communication within iGEM.
Read the full HumHub article, written by Stockholm and Alto-Helsinki, here
TU Eindhoven
Cloning methods represent one of the core elements of synthetic biology and it is crucial for each iGEM team to know as much as possible about the broad varity of cloning techniques. This need has been recognized by the iGEM Eindhoven which searched for collaborators using different cloning approaches to share their experiences in the latest cloning guide. The iGEM Team Stockholm could therefore contribute with their experience in overlap-extension PCR (OE-PCR) cloning. This technique is a completely restriction enzyme free approach to integrate elongated fragments into a target vector.