Difference between revisions of "Team:Goettingen/Experiments"
| (5 intermediate revisions by the same user not shown) | |||
| Line 37: | Line 37: | ||
} | } | ||
| − | #menu1, #menu2, #menu3, #menu4, #menu5, #menu6, #menu7, #menu8, #menu9, #menu10, #menu11, #menu12, #menu13, #menu14, #menu15, #menu16, #menu17, #menu18, #menu19, #menu20, #menu21, #menu22, #menu23, #menu24, #menu25, #menu26, #menu27, #menu28, #menu29, #menu30, #menu31, #menu32, #menu33, #menu34, #menu35, #menu36, #menu37, #menu38, #menu39, #menu40{ | + | #menu1, #menu2, #menu3, #menu4, #menu5, #menu6, #menu7, #menu8, #menu9, #menu10, #menu11, #menu12, #menu13, #menu14, #menu15, #menu16, #menu17, #menu18, #menu19, #menu20, #menu21, #menu22, #menu23, #menu24, #menu25, #menu26, #menu27, #menu28, #menu29, #menu30, #menu31, #menu32, #menu33, #menu34, #menu35, #menu36, #menu37, #menu38, #menu39, #menu40, #menu41{ |
display:none; | display:none; | ||
width:90%; | width:90%; | ||
| Line 206: | Line 206: | ||
</p> | </p> | ||
<p> | <p> | ||
| − | <a | + | <a href="https://www.dsmz.de/microorganisms/medium/pdf/DSMZ_Medium315.pdf">DSMZ Medium 315</a> |
</p> | </p> | ||
<p> | <p> | ||
| Line 212: | Line 212: | ||
</p> | </p> | ||
<p> | <p> | ||
| − | <a | + | <a href="https://www.dsmz.de/microorganisms/medium/pdf/DSMZ_Medium520.pdf"> DSMZ Medium 520</a> |
</p> | </p> | ||
<p> | <p> | ||
| Line 218: | Line 218: | ||
</p> | </p> | ||
<p> | <p> | ||
| − | <a | + | <a href="https://www.dsmz.de/microorganisms/medium/pdf/DSMZ_Medium122.pdf">DSMZ Medium 122</a> |
</p> | </p> | ||
<p> | <p> | ||
| Line 224: | Line 224: | ||
</p> | </p> | ||
<p> | <p> | ||
| − | <a | + | <a href="https://www.dsmz.de/microorganisms/medium/pdf/DSMZ_Medium165.pdf">DSMZ Medium 165</a> |
</p> | </p> | ||
<p> | <p> | ||
| − | <a | + | <a href="http://www.dsmz.de/microorganisms/medium/pdf/DSMZ_Medium141.pdf">DSMZ Medium 141</a> |
</p> | </p> | ||
| Line 865: | Line 865: | ||
<h2> Protein Extraction and Purification</h2> | <h2> Protein Extraction and Purification</h2> | ||
| + | <a href="" onClick=" $('#menu36').slideToggle(300, function callback() { }); return false;"><h1 style="color:white;"> Induction, harvest and disruption of expression cultures</h1></a> | ||
| + | <div id="menu36"> | ||
| + | <p> | ||
| + | <u>Induction</u> | ||
| + | </p> | ||
| + | <p> | ||
| + | Recombinant protein expression was exclusively realized in <em>E.coli</em>. pET101-ScaA was expressed in BL21(DE3) cells under the control of the <em>lac</em> operon, and pBAD-RFP-ACEL (and all other pBAD constructs) was expressed in TOP 10 cells under the control of the <em>ara</em> operon. The | ||
| + | procedure is as follows:<u></u> | ||
| + | </p> | ||
| + | <p> | ||
| + | Inoculate “fat” LB expression medium, having a volume corresponding to 5 % of the shake flask that is used, with 5 % (v/v) of an overnight LB starter | ||
| + | culture grown at 37°C and 150 rpm. | ||
| + | </p> | ||
| + | <p> | ||
| + | Incubate the expression culture at 37°C and 150 rpm until an OD<sub>600</sub> between 2.5 and 3.5 is reached. | ||
| + | </p> | ||
| + | <p> | ||
| + | Induce protein expression by adding the appropriate inducer to the expression culture. In case of pET101-ScaA use 1 mM IPTG and in case of pBAD-RFP-ACEL | ||
| + | use 0.2 % (v/v) L-arabinose. | ||
| + | </p> | ||
| + | <p> | ||
| + | Incubate the expression culture for 20 h at 37°C and 150 rpm. | ||
| + | </p> | ||
| + | <p> | ||
| + | Take a 1 ml sample before induction and before cell harvesting in order to analyze the expression effectivity of target proteins by SDS-PAGE. | ||
| + | </p> | ||
| + | <p> | ||
| + | <u>Cell Harvest</u> | ||
| + | </p> | ||
| + | <p> | ||
| + | Pellet the culture at 4°C, 13000rpm for 20min (SLA-3000 rotor, Sorvall). | ||
| + | </p> | ||
| + | <p> | ||
| + | Resuspend and wash with an equal volume of LEW Buffer (see Protein extraction). | ||
| + | </p> | ||
| + | <p> | ||
| + | Repeat previous centrifugation step. | ||
| + | </p> | ||
| + | <p> | ||
| + | Freeze pellet until further use or prepare for French Press. | ||
| + | </p> | ||
| + | <p> | ||
| + | <u>Cell extraction by French Press</u> | ||
| + | </p> | ||
| + | <p> | ||
| + | Resuspend pellet in 20mL 1x LEW Buffer . | ||
| + | </p> | ||
| + | <p> | ||
| + | Take a small sample (10µl) for microscopy. | ||
| + | </p> | ||
| + | <p> | ||
| + | Disrupt cells by using a discontinuous high-pressure homogenizer (e.g., French-Press, 4 cycles at 1000 psi). | ||
| + | </p> | ||
| + | <p> | ||
| + | take a small sample (10µl) and analyse both before and after press samples under the microscope. Look for inclusion bodies. | ||
| + | </p> | ||
| + | <p> | ||
| + | Sediment cell debris via centrifugation for 30 min at 13000 rpm (SS-34 rotor, Sorvall) and 4°C. | ||
| + | </p> | ||
| + | <p> | ||
| + | Filter the resulting supernatant through 0.45 µm sterile filters and subsequently apply it <a name="_GoBack"></a>to chromatographic purification steps. | ||
| + | </p> | ||
| + | </div> | ||
| + | |||
<a href="" onClick=" $('#menu29').slideToggle(300, function callback() { }); return false;"><h1 style="color:white;">Protein Purification (Protino® Ni-IDA 2000 His-Tag protein purification, Macherey-Nagel)</h1></a> | <a href="" onClick=" $('#menu29').slideToggle(300, function callback() { }); return false;"><h1 style="color:white;">Protein Purification (Protino® Ni-IDA 2000 His-Tag protein purification, Macherey-Nagel)</h1></a> | ||
<div id="menu29"> | <div id="menu29"> | ||
| Line 1,003: | Line 1,067: | ||
<p> | <p> | ||
Buffer B: Elution buffer (Lysis buffer supplemented with 250 mM imidazole) | Buffer B: Elution buffer (Lysis buffer supplemented with 250 mM imidazole) | ||
| + | </p> | ||
| + | |||
| + | </div> | ||
| + | <a href="" onClick=" $('#menu39').slideToggle(300, function callback() { }); return false;"><h1 style="color:white;"> Size Exclusion Chromatography</h1></a> | ||
| + | <div id="menu39"> | ||
| + | <p> | ||
| + | In a second purification step, the target proteins ScaA and RFP-ACEL were separated according to their size (radius of gyration) by employing size | ||
| + | exclusion chromatography. Therefore: | ||
| + | </p> | ||
| + | <p> | ||
| + | Connect a HiLoad Superdex 200 16/600 column (GE Healthcare) to an Äkta purifier or Äkta prime system and wash the column with 1 CV of 20 % EtOH and a | ||
| + | maximum flow rate of 1 ml/min. | ||
| + | </p> | ||
| + | <p> | ||
| + | Pre-equilibrate the column with loading buffer by washing first with 1 CV of water and subsequently with 1 CV of the respective buffer, each with a maximum | ||
| + | flow rate of 1 ml/min. | ||
| + | </p> | ||
| + | <p> | ||
| + | Load the concentrated protein solution with a maximum flow rate of 1 ml/min onto the column by using a 5 ml sampling loop. Start the automated | ||
| + | fractionation. | ||
| + | </p> | ||
| + | <p> | ||
| + | Elute the target protein with a maximum flow rate of 1 ml/min and analyze obtained peak fractions on a SDS gel. | ||
| + | </p> | ||
| + | <p> | ||
| + | Pool the fractions containing the target protein. | ||
| + | </p> | ||
| + | <p> | ||
| + | All chromatographic purification steps are carried out at room temperature. | ||
| + | </p> | ||
| + | <p> | ||
| + | Required buffer: | ||
| + | </p> | ||
| + | <p> | ||
| + | Buffer A: Loading buffer (20 mM phosphate pH 8, 500 mM NaCl, 5 % glycerol, 1 mM 2-mercaptoethanol) | ||
| + | </p> | ||
| + | |||
| + | </div> | ||
| + | <a href="" onClick=" $('#menu35').slideToggle(300, function callback() { }); return false;"><h1 style="color:white;"> Concentration of protein solutions</h1></a> | ||
| + | <div id="menu35"> | ||
| + | <p> | ||
| + | Concentration of protein solutions was realized via centrifugation using 20 ml Vivaspin centrifugal concentrators (Viva Science) with the appropriate | ||
| + | molecular weight cutoffs of 50 kDa in case of the scaffolding protein ScaA and 30 kDa in case of RFP-ACEL. In order to concentrate protein solutions: | ||
| + | </p> | ||
| + | <p> | ||
| + | Wash the concentrator with 20 ml of buffer in order to remove the protectant (glycerol) from the membrane. | ||
| + | </p> | ||
| + | <p> | ||
| + | Add the protein sample to the concentrator and start centrifugation at 4°C until the desired volume/concentration is reached. The sample volume and the | ||
| + | speed is determined by the rotor that is used. If a 25° fixed angle rotor is applied, the maximum starting volume is 14 ml and the speed is restricted to | ||
| + | 6000 x g. | ||
| + | </p> | ||
| + | <p> | ||
| + | Gently mix the upper reservoir prior to each refilling step and prior to transfer. | ||
| + | </p> | ||
| + | <p> | ||
| + | Check the flow through for leakage by Bradford assay. | ||
</p> | </p> | ||
| Line 1,038: | Line 1,159: | ||
</div> | </div> | ||
| + | <a href="" onClick=" $('#menu38').slideToggle(300, function callback() { }); return false;"><h1 style="color:white;"> SDS Polyacrylamid Gel Electrophoresis</h1></a> | ||
| + | <div id="menu38"> | ||
| + | <p> | ||
| + | The analytic protein separation according to the apparent molecular weight was carried out by discontinuous SDS polyacrylamid gel electrophoresis according | ||
| + | to Laemmli (1970) using mini gel electrophoresis chambers (BioRad, 10x8 cm). For cast and run of the gels do the following: | ||
| + | </p> | ||
| + | <p> | ||
| + | For the stacking gel use the following components: 0.25 % of the total volume of stacking gel buffer, and acrylamid to a final concentration of 4 %. Add | ||
| + | water to the final volume (cf. pipetting scheme). Add 1/133 of the total volume of 10 % ammonium persulfate (APS) and 1/1000 of | ||
| + | N,N,N’,N’-tetramethyl-ethylenediamine (TEMED). | ||
| + | </p> | ||
| + | <p> | ||
| + | For the separation gel mix the appropriate amount of acrylamid (e.g. 10 % for a 10 % SDS gel) with 0.25 % of the final volume of separation gel buffer and | ||
| + | fill with water to the final volume. Add 1/133 of the total volume of 10 % APS and 1/1000 of TEMED. | ||
| + | </p> | ||
| + | <p> | ||
| + | Place the poured gel into the running chamber and fill the upper and lower reservoir with running buffer. | ||
| + | </p> | ||
| + | <p> | ||
| + | Dilute the protein samples 1:1 with loading buffer and load the gel. | ||
| + | </p> | ||
| + | <p> | ||
| + | Run the gel with 15mA per gel for 15 min, and then increase to 30mA per Gel until the end. | ||
| + | </p> | ||
| + | <p> | ||
| + | After electrophoretic separation, place the gels into staining solution and heat it without boiling. | ||
| + | </p> | ||
| + | <p> | ||
| + | Incubate the gels under slow agitation until protein bands become visible. | ||
| + | </p> | ||
| + | <p> | ||
| + | Discolor the gels by using fixation solution. | ||
| + | </p> | ||
| + | <p> | ||
| + | <strong>Pipetting scheme. </strong> | ||
| + | </p> | ||
| + | <table border="1" cellspacing="0" cellpadding="0"> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | <strong>Compoment</strong> | ||
| + | </p> | ||
| + | <p align="center"> | ||
| + | <strong>(For 2 gels)</strong> | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | <strong>Stacking gel</strong> | ||
| + | </p> | ||
| + | <p align="center"> | ||
| + | <strong>(4 % acrylamid)</strong> | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | <strong>Separation gel</strong> | ||
| + | </p> | ||
| + | <p align="center"> | ||
| + | <strong>(10 % acrylamid)</strong> | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | Water | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | 2.64 ml | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | 4 ml | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | Acrylamid | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | 0.4 ml | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | 2 ml | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | Stacking gel buffer | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | 0.96 ml | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | --- | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | Separation gel buffer | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | --- | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | 2 ml | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="636" colspan="3" valign="top"> | ||
| + | <p align="center"> | ||
| + | <strong>Mix</strong> | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | 10 % APS | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | 30 µl | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | 60 µl | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | TEMED | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | 4 µl | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="212" valign="top"> | ||
| + | <p align="center"> | ||
| + | 6 µl | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | <p> | ||
| + | Required agents and solutions: | ||
| + | </p> | ||
| + | <p> | ||
| + | 40 % Acrylamid/bis-acrylamid (37.5:1) | ||
| + | </p> | ||
| + | <p> | ||
| + | 10 % (w/v) SDS solution | ||
| + | </p> | ||
| + | <p> | ||
| + | Stacking gel buffer (1.5 M Tris/HCl pH 8.8, 0.4 % (w/v) SDS) | ||
| + | </p> | ||
| + | <p> | ||
| + | Separation gel buffer (0.5 M Tris/HCl pH 6.5, 0.4 % (w/v) SDS) | ||
| + | </p> | ||
| + | <p> | ||
| + | 2x Laemmli loading buffer (20 mM Tris/HCl pH 6.5, 4 % (w/v) SDS, 10 % (v/v) β-mercaptoethanol, 40 % (v/v) glycerol, 0.002 % (w/v) bromphenol blue) | ||
| + | </p> | ||
| + | <p> | ||
| + | 1x SDS running buffer (25 mM Tris base, 192 mM glycine, 0.1 % (w/v) SDS) | ||
| + | </p> | ||
| + | <p> | ||
| + | Staining solution (0.005 % (w/v) Coomassie Brilliant Blue G-250, 0.0025 % (w/v) Coomassie Brilliant Blue R-250, 10 % (v/v) ethanol und 5 % (v/v) acetic | ||
| + | acid) | ||
| + | </p> | ||
| + | <p> | ||
| + | Fixation solution (10 % (v/v) acetic acid) | ||
| + | </p> | ||
| + | </div> | ||
| − | <h2> Activity Screens </h2> | + | <a href="" onClick=" $('#menu40').slideToggle(300, function callback() { }); return false;"><h1 style="color:white;"> Western Blot</h1></a> |
| + | <div id="menu40"> | ||
| + | <p> | ||
| + | 10 x TBS Buffer (store at RT) | ||
| + | </p> | ||
| + | <table border="1" cellspacing="0" cellpadding="0"> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | Tris HCL (MW: 121,14 g/mol) | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | 6.057g | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | NaCl (MW: 58.44 g/mol) | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | 40.91g | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | H<sub>2</sub>O dest. | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | ad. 500 ml | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | <p> | ||
| + | adjust to pH 7.4 | ||
| + | </p> | ||
| + | <p> | ||
| + | 1 x Transfer Buffer (store at RT) | ||
| + | </p> | ||
| + | <table border="1" cellspacing="0" cellpadding="0"> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | Tris | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | 3.029 g | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | Glycin | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | 14,41g | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | Methanol | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | 200ml | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | H<sub>2</sub>O bidest. | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | ad. 1000ml | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | <p> | ||
| + | check pH: 8.1-8.4 | ||
| + | </p> | ||
| + | <p> | ||
| + | If the pH is not correct, the Buffer has to be remade. Do not adjust. | ||
| + | </p> | ||
| + | <p> | ||
| + | 10 x TBS-Tween Buffer (store at RT) | ||
| + | </p> | ||
| + | <table border="1" cellspacing="0" cellpadding="0"> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | Tris-HCL (MW: 121.14 g/mol) | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | 6.057g | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | NaCl (MW: 58.44 g/mol) | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | 40.91g | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | Tween20 (Endkonz. 0,1%) | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | 0.5ml | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | H<sub>2</sub>O bidest. | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | ad. 500ml | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | <p> | ||
| + | adjust to pH 7.4 | ||
| + | </p> | ||
| + | <p> | ||
| + | Blocking Solution | ||
| + | </p> | ||
| + | <table border="1" cellspacing="0" cellpadding="0"> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | Skim Milk | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | 2,5g | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | 1 x TBS | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | 50ml | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | <p> | ||
| + | Dissolve for 15-30 min at RE. Store 400µl at 4°C for a later step. | ||
| + | </p> | ||
| + | <p> | ||
| + | Phosphatase Buffer (store at RT) | ||
| + | </p> | ||
| + | <table border="1" cellspacing="0" cellpadding="0"> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | Tris-HCL (MW: 121.14 g/mol) | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | 6.057g | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | NaCl (MW: 58.44 g/mol) | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | 2.922 g | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | MgCl<sub>2 </sub>x 6 H<sub>2</sub>O (MW: 203.3 g/mol) | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | 0.51 g | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | H<sub>2</sub>O bidest. | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="307" valign="top"> | ||
| + | <p> | ||
| + | ad. 500ml | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | <p> | ||
| + | adjust to pH 9.5 | ||
| + | </p> | ||
| + | <p> | ||
| + | Antibody: Anti V5-AP Antibody (Invitrogen, store at 4°C) | ||
| + | </p> | ||
| + | <p> | ||
| + | NBT: Promega, 50 mg/ml, store at -20°C | ||
| + | </p> | ||
| + | <p> | ||
| + | BCIP: Promega, 50 mg/ml, store at -20°C | ||
| + | </p> | ||
| + | <p> | ||
| + | 1.) Run an SDS Gel and use Protein Ladder compatible with Western Blots. | ||
| + | </p> | ||
| + | <p> | ||
| + | 2.) Use Perfect Blue Semi-Dry Electro blotter from Peqlab and assemble apparatus in a tip box with a little transfer buffer (sandwich should be moist <u>not </u>wet): | ||
| + | </p> | ||
| + | <p> | ||
| + | Base of blotter = Kathode (+) bottom | ||
| + | </p> | ||
| + | <p> | ||
| + | 3x Whatman paper (moist) size of SDS page | ||
| + | </p> | ||
| + | <p> | ||
| + | Blotting membrane (Hybond EHL, 0,45µm, moist, no bubbles, SDS page sized) | ||
| + | </p> | ||
| + | <p> | ||
| + | SDS Gel (no bubbles! Proteins have a neg. charge and go towards the Kathode) | ||
| + | </p> | ||
| + | <p> | ||
| + | 3x Whatman paper (moist, gel sized) | ||
| + | </p> | ||
| + | <p> | ||
| + | Lid = Anode (-) top | ||
| + | </p> | ||
| + | <p> | ||
| + | After assembly of the sandwich the lid is screwed on lightly with 3 screws. Connect Blotter to an adequate power supply, which can work at a constant 10V. | ||
| + | Blot at 200mA per Gel and constant 10V for 1 hour. | ||
| + | </p> | ||
| + | <p> | ||
| + | 3.) Wash the membrane 3x 5 min with 1x TBS Buffer on a rocker | ||
| + | </p> | ||
| + | <p> | ||
| + | 4.) Block membrane in 50ml 1 x TBS Buffer with 5% skim milk for one hour or overnight on a rocker. | ||
| + | </p> | ||
| + | <p> | ||
| + | 5.) Wash 3x 15min 1xTBS-Tween on a rocker | ||
| + | </p> | ||
| + | <p> | ||
| + | 6.) Prepare Antibody: 14ml 1xTBS-Tween + 200µl 1x TBS Puffer/5% Skimmilk + 7µl Antibody | ||
| + | </p> | ||
| + | <p> | ||
| + | 7.) Incubate with antibody for 2 hours on a rocker | ||
| + | </p> | ||
| + | <p> | ||
| + | 8.) Wash 2x shortly in 1x TBS-Tween Buffer on a rocker | ||
| + | </p> | ||
| + | <p> | ||
| + | 9.) 1x 15min 1x TBS-Tween Buffer on the rocker | ||
| + | </p> | ||
| + | <p> | ||
| + | 10.) 2x 5min 1x TBS-Tween Buffer on the rocker | ||
| + | </p> | ||
| + | <p> | ||
| + | 11.) Wash 2x 10 min 1x TBS Buffer on a rocker | ||
| + | </p> | ||
| + | <p> | ||
| + | 12.) Wash 2x shortly in 1x Phosphatse buffer | ||
| + | </p> | ||
| + | <p> | ||
| + | 13.) 10ml 1x Phosphatase buffer + 66µl NBT + 33µl BCIP | ||
| + | </p> | ||
| + | <p> | ||
| + | - stain in the dark until bands become visible | ||
| + | </p> | ||
| + | <p> | ||
| + | - stop the reaction by washing with H2O | ||
| + | </p> | ||
| + | <p> | ||
| + | - dry western blot in Whatman Paper | ||
| + | </p> | ||
| + | <p> | ||
| + | 14.) Scan the membrane | ||
| + | </p> | ||
| + | |||
| + | </div> | ||
| + | <h2> Activity Screens & Tests</h2> | ||
<a href="" onClick=" $('#menu9').slideToggle(400, function callback() { }); return false;"><h1>Esterase activity test</h1></a> | <a href="" onClick=" $('#menu9').slideToggle(400, function callback() { }); return false;"><h1>Esterase activity test</h1></a> | ||
<div id="menu9"> | <div id="menu9"> | ||
| Line 1,092: | Line 1,760: | ||
<li>Incubate plates at 37<sup>o</sup>C for minimum 3-4 days (timing can be extended depending on bacterial strain).</li> | <li>Incubate plates at 37<sup>o</sup>C for minimum 3-4 days (timing can be extended depending on bacterial strain).</li> | ||
<li>After 3-4 days look for plates having clear zone of activity around cellulose substrate.</li> | <li>After 3-4 days look for plates having clear zone of activity around cellulose substrate.</li> | ||
| + | </div> | ||
| + | |||
| + | <a href="" onClick=" $('#menu41').slideToggle(300, function callback() { }); return false;"><h1 style="color:white;"> Enzymatic Activity Test for Cellulase </h1></a> | ||
| + | <div id="menu41"> | ||
| + | |||
| + | |||
| + | <p> | ||
| + | Solutions: | ||
| + | </p> | ||
| + | <p> | ||
| + | Buffer: | ||
| + | </p> | ||
| + | <p> | ||
| + | Tris (MW: 121.14 g/mol) 50 mM | ||
| + | </p> | ||
| + | <p> | ||
| + | dH<sub>2</sub>O ad 1 l | ||
| + | </p> | ||
| + | <p> | ||
| + | pH 7 | ||
| + | </p> | ||
| + | <p> | ||
| + | Substrate solution: | ||
| + | </p> | ||
| + | <p> | ||
| + | Carboxymethil cellulose 1 % (w / v) | ||
| + | </p> | ||
| + | <p> | ||
| + | ddH<sub>2</sub>O ad 1 l | ||
| + | </p> | ||
| + | <p> | ||
| + | DNSA reagent solution: | ||
| + | </p> | ||
| + | <p> | ||
| + | Dinitrosalycylic acid 10 g | ||
| + | </p> | ||
| + | <p> | ||
| + | Phenol 2 ml | ||
| + | </p> | ||
| + | <p> | ||
| + | K Na Tartrate 200 g | ||
| + | </p> | ||
| + | <p> | ||
| + | NaOH 10 g | ||
| + | </p> | ||
| + | <p> | ||
| + | Na<sub>2</sub>SO<sub>3</sub> 0.5 g | ||
| + | </p> | ||
| + | <p> | ||
| + | dH<sub>2</sub>O ad 1 l | ||
| + | </p> | ||
| + | <p> | ||
| + | Store at 4°C, protected from light | ||
| + | </p> | ||
| + | <p> | ||
| + | Procedure: | ||
| + | </p> | ||
| + | <p> | ||
| + | 250 µl substrate solution | ||
| + | </p> | ||
| + | <p> | ||
| + | 100 µl Tris-Buffer | ||
| + | </p> | ||
| + | <p> | ||
| + | x µl enzyme solution | ||
| + | </p> | ||
| + | <p> | ||
| + | ad 500 µl ddH<sub>2</sub>O | ||
| + | </p> | ||
| + | <p> | ||
| + | Incubate setup as described above at 37°C for 20 min. | ||
| + | </p> | ||
| + | <p> | ||
| + | Add 750 µl DNSA. Heat the setup at 96°C for 15 min. | ||
| + | </p> | ||
| + | <p> | ||
| + | Put mixture on ice, centrifuge shortly at 4°C, 13000 x g. | ||
| + | </p> | ||
| + | <p> | ||
| + | Measure OD<sub>575</sub> against same setup without enzyme as a blank. | ||
| + | </p> | ||
</div> | </div> | ||
Latest revision as of 00:00, 19 September 2015
Media/Buffer
LB Medium
"Fat" LB Medium
Media and Culture Methods for Dockerin Organisms of Origin
Phosphatase Activity plates, Sperber media
Esterase Activity plates, with 1% Tributyrin
Cellulase activity plates
1x TAE Buffer
Cloning Methods
PCR product purification using QIAquick® PCR Purification Kit (QIAGEN)
PCR Gel extraction, peqGOLD Gel Extraction Kit
Blunt End Ligation in pJET1.2 vector –Clone JET PCR Cloning Kit– (Thermo Scientific)
Sticky End T4 Ligation (Thermo Scientific)
TOPO® Cloning protocol usingChampion™ pET Directional TOPO® Expression Kits (Thermo Fisher Scientific)
Plasmid transformation into chemically competent E. coli
Electroporation of BL21 cells with pJET_RFP
Plasmid Extraction - using QIAprep Spin Miniprep Kit (QIAGEN)
Plasmid Extraction - using peqGOLD Plasmid Miniprep Kit I (PEQLAB Technologies)
Competent Cells
Preparation of competent E.coli cells
Transformation Efficiency Kit, RFP construct (iGEM)
Protein Extraction and Purification
Induction, harvest and disruption of expression cultures
Protein Purification (Protino® Ni-IDA 2000 His-Tag protein purification, Macherey-Nagel)
Affinity chromatography of His-tagged proteins
Size Exclusion Chromatography
Concentration of protein solutions
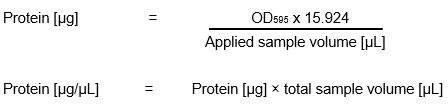
Bradford Assay
SDS Polyacrylamid Gel Electrophoresis
Western Blot
Activity Screens & Tests
Esterase activity test
Phosphatase activity test
Cellulase activity screening
Enzymatic Activity Test for Cellulase
Restriction Controls
Aan I (Psi I ) - thermo fisher scientific - restriction control protocol
Double digestion restriction control
Restriction control using fast and slow digestion enzymes
Scafoldin Restriction control
Esterase Restriction Control
Phosphatase Restriction Control
PCR Preparation Methods
Colony PCR
Phusion PCR
Sequencing
Protocol for Sanger sequencing
Overnight Sanger Sequencing
Fluorescence Microscopy
RFP microscopy