Difference between revisions of "Team:Goettingen/Results"
| Line 39: | Line 39: | ||
} | } | ||
| − | #menu1, #menu2, #menu3{ | + | #menu1, #menu2, #menu3, #menu4{ |
display:none; | display:none; | ||
width:90%; | width:90%; | ||
| Line 365: | Line 365: | ||
| − | < | + | <a href="" onClick=" $('#menu4').slideToggle(300, function callback() { }); return false;"><h1>BioBricks</h1></a> |
| − | + | <div id="menu4"> | |
| − | + | ||
| − | < | + | |
| − | + | ||
| − | + | ||
| − | < | + | |
| − | + | ||
| − | <p> | + | <p> |
| + | <strong>Dockerin Biobricks</strong> | ||
| + | </p> | ||
| + | <p> | ||
| + | The dockerins ACEL (<em>Acetivibrio cellulolyticus</em>), BCEL (<em>Bacteroides cellulosolvens</em>), CCEL (<em>Clostridium cellulolyticum</em>) and CTHE (<em>Clostridium thermocellum</em>) were amplified from pBAD_ACEL, pBAD_BCEL, pBAD_CCEL and pBAD_CTHE with primers to add the desired <em>Eco</em>RI and <em>Pst</em>I restriction sites by PCR. Primers contained restriction sites for <em>Eco</em>RI and <em>Pst</em>I in order to make them compatible for | ||
| + | insertion into the iGEM shipping vector pSB1C3. | ||
| + | </p> | ||
| + | <p> | ||
| + | After purification, the PCR products were restricted with <em>Eco</em>RI and <em>Pst</em>I, as well as pSB1C3. Afterwards the restricted dockerins were | ||
| + | ligated into pSB1C3 by T4 ligation (sticky end ligation) and transformed into <em>E. coli</em> TOP10. | ||
| + | </p> | ||
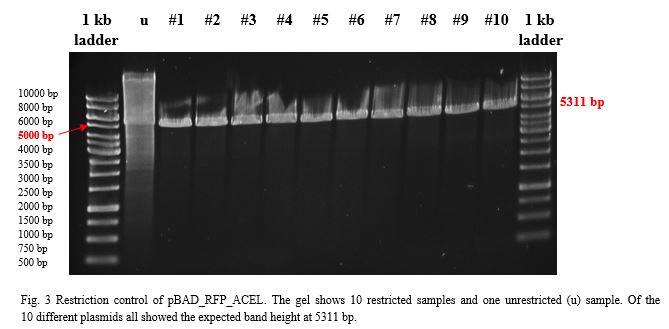
| + | <p> | ||
| + | Ligation into pSB1C3 was followed according to the protocol in the methods collection. After over-night incubation colonies were picked, plasmids extracted | ||
| + | with the QIAGEN QIAprep Spin Miniprep Kit and test restricted with <em>Eco</em>RI (pSB1C3_CCEL and pSB1C3_ACEL) and <em>Eco</em>RI and <em>Pst</em>I | ||
| + | (pSB1C3_BCEL) (Fig.1). We lost CTHE due to failing transformations at this point. | ||
| + | </p> | ||
| − | + | </html> | |
| − | + | [[File:Rest_control_pSB1C3_ACEL_CCEL_BCEL.jpg|600px|thumb|center|]] | |
| − | + | <html> | |
| − | < | + | |
| + | <p> | ||
| + | Once restriction controls showed the correct bands, pSB1C3_CCEL, pSB1C3_ACEL and pSB1C3_BCEL were sent for sequencing. | ||
| + | </p> | ||
| + | <p> | ||
| + | <strong>Colour BioBricks</strong> | ||
| + | </p> | ||
| + | <p> | ||
| + | We also wanted to improve already existing BioBricks by fusing our dockerins to the colours eforRed (BBa_K592012) and amilCP (BBa_K592009) that were | ||
| + | submitted from the University of Uppsala (Sweden) in 2011. | ||
| + | </p> | ||
| + | <p> | ||
| + | So we chose to fuse eforRed to BCEL and amilCP to CCEL. Since the plasmids of the two colour proteins were not distributed with the current plate of the | ||
| + | iGEM Distribution Kit, we ordered those parts including the right restriction sites as gBlocks from IDT. | ||
| + | </p> | ||
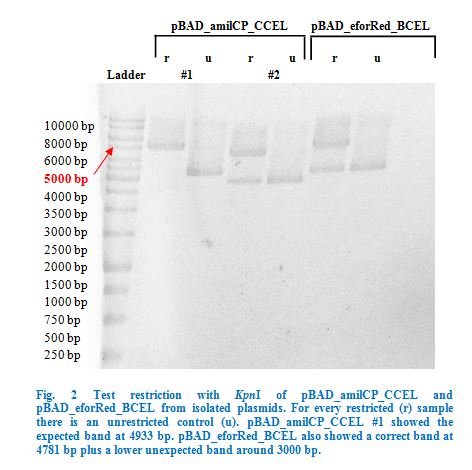
| + | <p> | ||
| + | As a first step eforRed and amilCP were restricted with <em>Kpn</em>I and <em>Sac</em>I to make them compatible with our dockerins in pBAD. | ||
| + | </p> | ||
| + | <p> | ||
| + | After purification restricted colours were ligated into pBAD_BCEL and pBAD_CCEL by T4 ligation (sticky end ligation) and transformed into <em>E. coli</em> | ||
| + | TOP10. | ||
| + | </p> | ||
| + | <p> | ||
| + | Ligation into pBAD was followed according to the protocol in the methods collection. After over-night incubation colonies were picked, plasmids extracted | ||
| + | with the QIAGEN QIAprep Spin Miniprep Kit and test restricted with <em>Kpn</em>I (Fig.2). | ||
| + | </p> | ||
| + | </html> | ||
| + | [[File:TeamGoettingen2015_Rest_control_pBAD_eforRed_BCEL_and_amilCP_CCEL_Goettingen2015.jpeg|600px|thumb|center|]] | ||
| + | <html> | ||
| − | < | + | <p> |
| − | <p> | + | Once restriction controls showed the correct bands, pBAD_amilCP_CCEL #1 and pBAD_eforRed_BCEL were sent for sequencing. Sequencing showed that both |
| − | < | + | plasmids were correct. |
| − | < | + | </p> |
| − | < | + | <p> |
| − | < | + | To make our constructs compatible with the iGEM shipping vector pSB1C3 the desired <em>Eco</em>RI and <em>Pst</em>I restrictions sites were added by PCR. |
| − | </ | + | </p> |
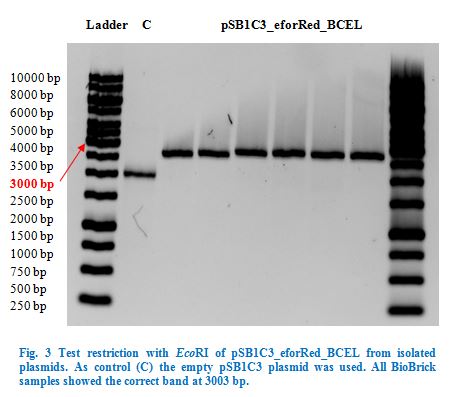
| + | <p> | ||
| + | After purification, the PCR products were restricted with <em>Eco</em>RI and <em>Pst</em>I, the same with pSB1C3, and afterwards ligated into pSB1C3 by T4 | ||
| + | ligation (sticky end ligation) and transformed into <em>E. coli</em> TOP10. | ||
| + | </p> | ||
| + | <p> | ||
| + | After over-night incubation colonies were picked, plasmids extracted with the QIAGEN QIAprep Spin Miniprep Kit and test restricted with <em>Eco</em>RI | ||
| + | (Fig.3). Due to the reason that our dockerin CCEL contains an internal <em>Pst</em>I restriction site we continued working only with the eforRed_BCEL | ||
| + | construct. | ||
| + | </p> | ||
| + | |||
| + | </html> | ||
| + | [[File:TeamGoettingen2015_Rest_control_pSB1C3_eforRed_BCEL_Goettingen2015.jpeg|600px|thumb|center|]] | ||
| + | <html> | ||
| + | |||
| + | <p> | ||
| + | Once restriction controls showed the correct band, pSB1C3_eforRed_BCEL was sent for sequencing. Sequencing showed that the plasmids were correct. | ||
| + | </p> | ||
| + | <p> | ||
| + | <strong>RESULTS</strong> | ||
| + | </p> | ||
| + | <p> | ||
| + | Sequencing showed that the dockerins ACEL and CCEL contain an internal <em>Pst</em>I restriction site. Therefore all the constructs containing ACEL or CCEL | ||
| + | showed in the end truncated dockerin sequences and could not be send in as BioBricks. | ||
| + | </p> | ||
| + | <p> | ||
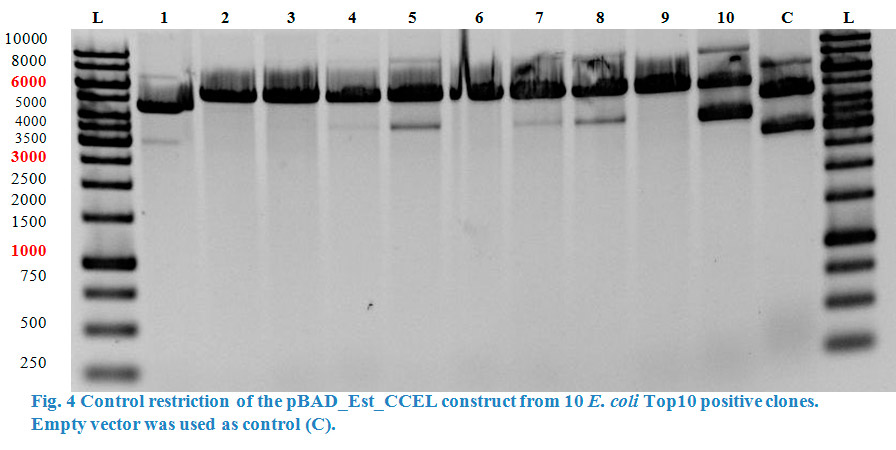
| + | We also build a construct where the enzyme esterase was fused to our CCEL dockerin (pBAD_Est_CCEL) following the same strategy. But again, due to the | ||
| + | reason that our dockerin CCEL contains an internal <em>Pst</em>I restriction site we had to stop our work at this point. | ||
| + | </p> | ||
| + | <p> | ||
| + | Furthermore we collaborated with the current iGEM team of Aachen and tried to fuse three of their enzymes to our constructs (see Collaboration link) but | ||
| + | could not finish our work here. | ||
| + | </p> | ||
| + | <p> | ||
| + | <strong>ACHIEVENTS<a name="_GoBack"></a></strong> | ||
| + | </p> | ||
| + | <p> | ||
| + | <strong>Sequences of pSB1C3_BCEL and pSB1C3_eforRed_BCEL were correct and submitted as BioBricks to iGEM:</strong> | ||
| + | </p> | ||
| + | <p> | ||
| + | pSB1C3_BCEL (BBa_1865000) | ||
| + | </p> | ||
| + | <p> | ||
| + | pSB1C3_eforRed_BCEL (BBa_1865001) | ||
| + | </p> | ||
</div> | </div> | ||
</html> | </html> | ||
Revision as of 18:06, 18 September 2015