Difference between revisions of "Team:Austin UTexas/Project/Plasmid Study"
Kvasudevan (Talk | contribs) |
Kvasudevan (Talk | contribs) m |
||
| Line 45: | Line 45: | ||
<font face="Courier New">Protocol</font> | <font face="Courier New">Protocol</font> | ||
<hr> | <hr> | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
<br> | <br> | ||
<br> | <br> | ||
<br> | <br> | ||
Revision as of 19:14, 11 September 2015
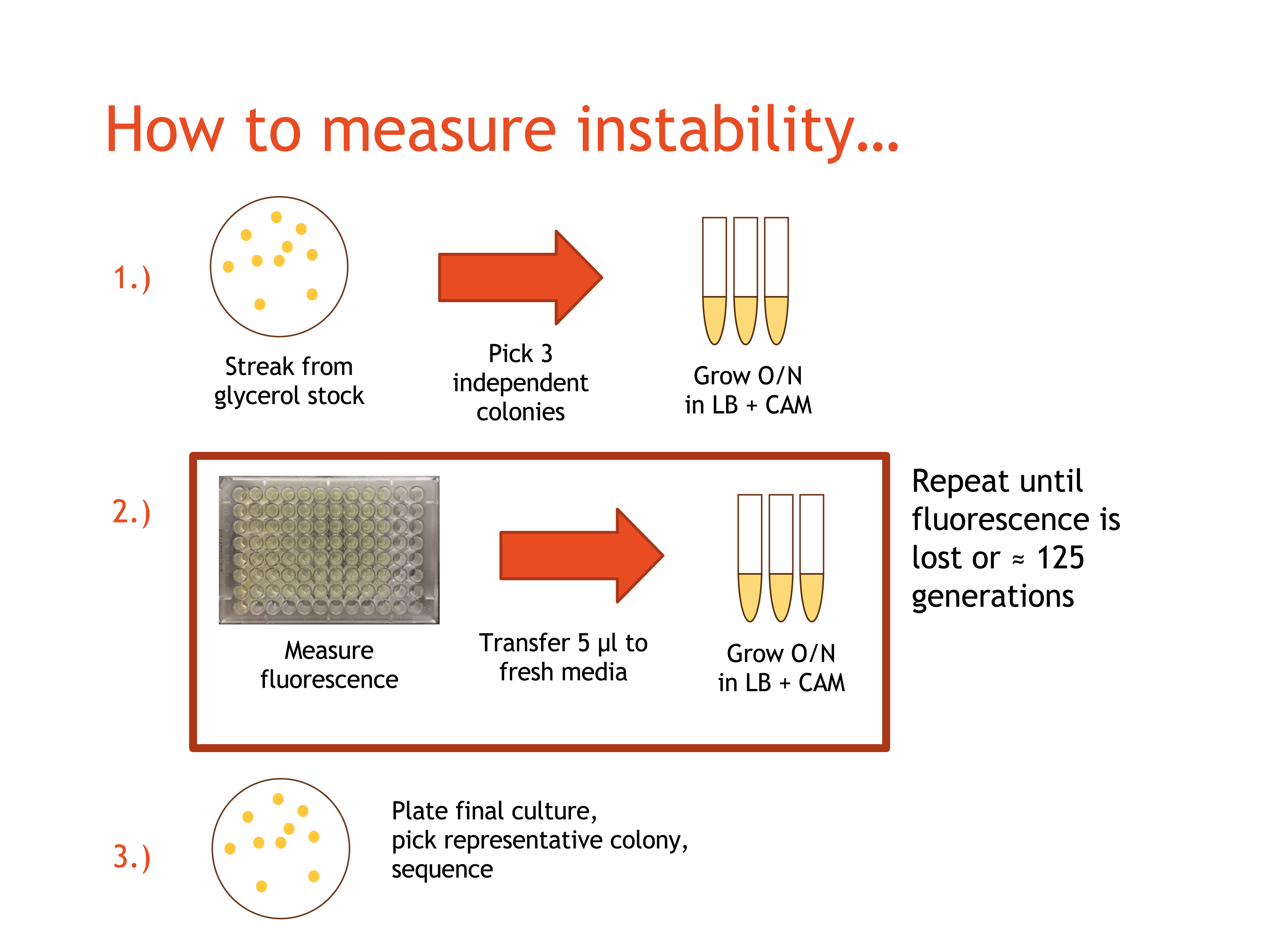
--this is a first draft--
Spring 2015 Data Set
Plasmid Construction and Transformation
Plasmids were constructed with standard BioBrick cloning procedures and enzymes. Composite plasmids were constructed using the following BioBrick combinations:
REPLACE LIST WITH TABLE AND/OR FIGURE
BBa_K864100+BBa_K314100BBa_E0020+BBa_K608007
BBa_EE030+BBa_K608006
BBa_E0020+BBa_K608003
BBa_E0030+BBa_K608007
BBa_K592101+BBa_K608006
BBa_K592100+BBa_K608004
BBa_K592101+BBa_K608002
BBa_K864100+BBa_K608006
BBa_E0030+BBa_K314100
BBa_E0020+BBa_K608002
BBa_K864100+BBa_K608002
BBa_E0030+BBa_K314100
BBa_K592101+BBa_K608007