Difference between revisions of "Team:Austin UTexas/Project/Plasmid Study"
Kvasudevan (Talk | contribs) m |
Kvasudevan (Talk | contribs) |
||
| Line 1: | Line 1: | ||
{{Austin_UTexas}} | {{Austin_UTexas}} | ||
| − | <font | + | <font size="5">Spring Protocol</font> |
| + | <hr> | ||
<font size="3"> | <font size="3"> | ||
<br> | <br> | ||
| Line 128: | Line 129: | ||
<div align=left> | <div align=left> | ||
<br> | <br> | ||
| − | Several fluorescent protein/promoter+RBS combinations were studied in more than one experiment, which is why each is classified as a 'unique' combination.<br> | + | Several fluorescent protein/promoter+RBS combinations were studied in more than one experiment, which is why each plasmid above is classified as a 'unique' combination.<br> |
<br> | <br> | ||
The BioBricks in each pair were ligated together using T4 Ligase, and transformed into TOP10 <i>E. coli</i>. Prepared cultures were streaked onto LB/chloramphenicol (CAM) agar plates, and were placed in a 37°C incubator overnight. After at least 20 hours, the plates were retrieved,and the one exhibiting most fluorescent colonies and for each team member participating in the experiment was selected. Three independent, fluorescent colonies were selected and each placed into 5mL of LB/CAM media. The tubes for each were labelled with the date, the culture name, DAY 1, and the strain used. The tubes were placed in a 37°C shaker-incubator and the plates were placed in a 4°C cold room for storage. | The BioBricks in each pair were ligated together using T4 Ligase, and transformed into TOP10 <i>E. coli</i>. Prepared cultures were streaked onto LB/chloramphenicol (CAM) agar plates, and were placed in a 37°C incubator overnight. After at least 20 hours, the plates were retrieved,and the one exhibiting most fluorescent colonies and for each team member participating in the experiment was selected. Three independent, fluorescent colonies were selected and each placed into 5mL of LB/CAM media. The tubes for each were labelled with the date, the culture name, DAY 1, and the strain used. The tubes were placed in a 37°C shaker-incubator and the plates were placed in a 4°C cold room for storage. | ||
| Line 139: | Line 140: | ||
<br> | <br> | ||
<br> | <br> | ||
| − | The | + | The primary aim of this project was to assess the fluorescent protein coding sequences that are most prone to breaking/mutating. In order to do this, we had to propagate different fluorescent proteins and record their fluorescence daily. |
| + | <br> | ||
| + | <br> | ||
| + | The day immediately following the start of the DAY 1 cultures, the DAY 1 cultures were retrieved and resuspendeded. Three empty culture tubes for each participating team member were collected and labelled the same way as the first set of culture tubes, but with DAY 2 instead of DAY 1. 5mL of LB/CAM media were placed into each culture tube, and 5μL of culture from the microcentrifuge tubes were pipetted into the appropriate culture tube. The culture tubes for DAY 2 were placed into the shaker-incubator, and the DAY 1 culture tubes were placed in the cold room. <br> | ||
<br> | <br> | ||
<p>200μL of each DAY 1 culture were pipetted into a well on a 96-well plate (which was stored on ice when not in use). The well number, date, and culture name were recorded, and the plates were read for fluorescence the following morning. This process of moving cultures forward and obtaining fluorescence readings was repeated daily until the culture fluorescence dropped dramatically, or until DAY 10, so that each culture had a maximum 10 culture tubes over the 10 days.</p><br> | <p>200μL of each DAY 1 culture were pipetted into a well on a 96-well plate (which was stored on ice when not in use). The well number, date, and culture name were recorded, and the plates were read for fluorescence the following morning. This process of moving cultures forward and obtaining fluorescence readings was repeated daily until the culture fluorescence dropped dramatically, or until DAY 10, so that each culture had a maximum 10 culture tubes over the 10 days.</p><br> | ||
| Line 148: | Line 152: | ||
<br> | <br> | ||
<br> | <br> | ||
| + | <font size="5">Summer Protocol</font> | ||
| + | <hr> | ||
| + | <hr> | ||
| + | |||
| + | |||
<br> | <br> | ||
Revision as of 23:58, 13 September 2015
Spring Protocol
Spring 2015 Data Set
Plasmid Construction and Transformation
Plasmids were constructed with standard BioBrick cloning procedures and enzymes. Composite plasmids were constructed using the following unique BioBrick combinations:
| Plasmid | Fluorescent Protein BioBrick | Fluorescent Protein | Promoter/RBS BioBrick | Promoter;RBS Strength |
| 1 | BBa_K592100 | BFP | BBa_K608004 | Strong; Weak |
| 2 | BBa_K592100 | BFP | BBa_K608007 | Medium; Weak |
| 3 | BBa_E0020 | ECFP | BBa_K608007 | Medium; Weak |
| 4 | BBa_E0020 | ECFP | BBa_K608003 | Strong; Medium |
| 5 | BBa_E0020 | ECFP | BBa_K608002 | Strong; Strong |
| 6 | BBa_K592101 | YFP | BBa_K608002 | Strong; Strong |
| 7 | BBa_K592101 | YFP | BBa_K608006 | Medium; Medium |
| 8 | BBa_K592101 | YFP | BBa_K608007 | Medium; Weak |
| 9 | BBa_K864100 | SYFP2 | BBa_K608002 | Strong; Strong |
| 10 | BBa_K864100 | SYFP2 | BBa_K314100 | Strong; Very Strong |
| 11 | BBa_K864100 | SYFP2 | BBa_K608006 | Medium; Medium |
| 12 | BBa_E0030 | EYFP | BBa_K608007 | Medium; Weak |
| 13 | BBa_EE030 | EYFP | BBa_K608006 | Medium; Medium |
| 14 | BBa_E0030 | EYFP | BBa_K314100 | Strong; Very Strong |
Several fluorescent protein/promoter+RBS combinations were studied in more than one experiment, which is why each plasmid above is classified as a 'unique' combination.
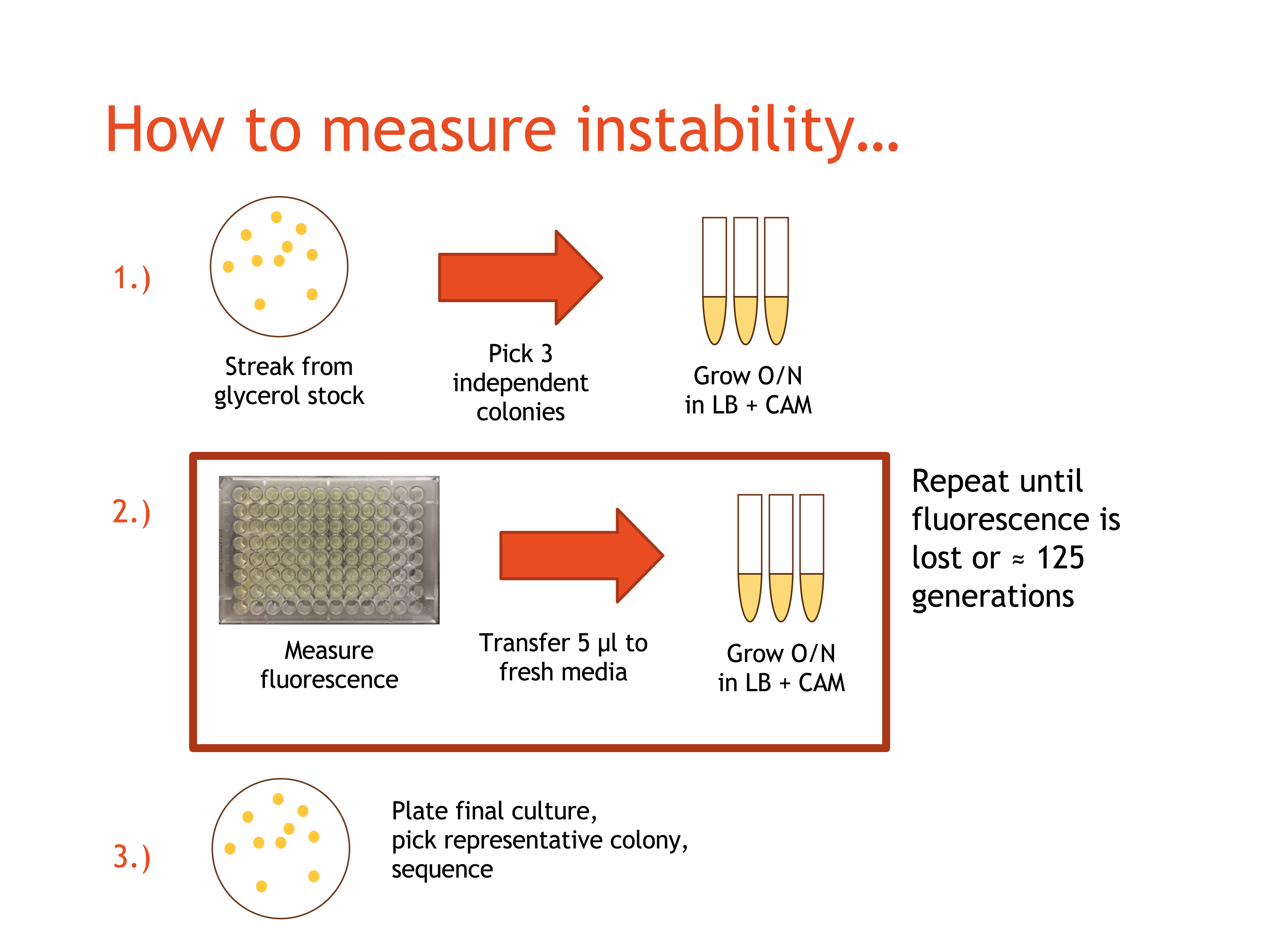
The BioBricks in each pair were ligated together using T4 Ligase, and transformed into TOP10 E. coli. Prepared cultures were streaked onto LB/chloramphenicol (CAM) agar plates, and were placed in a 37°C incubator overnight. After at least 20 hours, the plates were retrieved,and the one exhibiting most fluorescent colonies and for each team member participating in the experiment was selected. Three independent, fluorescent colonies were selected and each placed into 5mL of LB/CAM media. The tubes for each were labelled with the date, the culture name, DAY 1, and the strain used. The tubes were placed in a 37°C shaker-incubator and the plates were placed in a 4°C cold room for storage.
Fluorescence Readings
The primary aim of this project was to assess the fluorescent protein coding sequences that are most prone to breaking/mutating. In order to do this, we had to propagate different fluorescent proteins and record their fluorescence daily.
The day immediately following the start of the DAY 1 cultures, the DAY 1 cultures were retrieved and resuspendeded. Three empty culture tubes for each participating team member were collected and labelled the same way as the first set of culture tubes, but with DAY 2 instead of DAY 1. 5mL of LB/CAM media were placed into each culture tube, and 5μL of culture from the microcentrifuge tubes were pipetted into the appropriate culture tube. The culture tubes for DAY 2 were placed into the shaker-incubator, and the DAY 1 culture tubes were placed in the cold room.
200μL of each DAY 1 culture were pipetted into a well on a 96-well plate (which was stored on ice when not in use). The well number, date, and culture name were recorded, and the plates were read for fluorescence the following morning. This process of moving cultures forward and obtaining fluorescence readings was repeated daily until the culture fluorescence dropped dramatically, or until DAY 10, so that each culture had a maximum 10 culture tubes over the 10 days.
The DAY 10 cultures were allowed to incubate and were sent for sequencing. A full "control" plasmid sequence was created for each culture by inserting the recorded BioBrick sequences into an existing pSB1C3-circular sequence on Benchling.com. The sequencing results were uploaded to Benchling and compared to the predicted original sequence of the plasmid for any possible mutations.
summary figure(?)
Summer Protocol