Difference between revisions of "Team:Goettingen/Results"
| Line 256: | Line 256: | ||
<p>After purification of the PCR product it was ligated into pJET1.2 by subcloning (blunt end ligation). This vector serves to clone the fluorescent protein without triggering its activity, which may interact with the expression vector or the chosen <em>E.coli </em>strain.</p> | <p>After purification of the PCR product it was ligated into pJET1.2 by subcloning (blunt end ligation). This vector serves to clone the fluorescent protein without triggering its activity, which may interact with the expression vector or the chosen <em>E.coli </em>strain.</p> | ||
<p>Ligation into pJET1.2 was followed according to the protocol in the methods collection. After over-night incubation colonies were picked, plasmids extracted with the QIAGEN QIAprep Spin Miniprep Kit and restricted with <em>Kpn</em>I and <em>Sac</em>I.</p> | <p>Ligation into pJET1.2 was followed according to the protocol in the methods collection. After over-night incubation colonies were picked, plasmids extracted with the QIAGEN QIAprep Spin Miniprep Kit and restricted with <em>Kpn</em>I and <em>Sac</em>I.</p> | ||
| + | <p>Once restriction controls showed the correct bands, both pJET_RFP_3 and pJET_RFP_7 were sent for sequencing by the G2L laboratory.</p> | ||
<img src=" https://static.igem.org/mediawiki/2015/1/1d/Restr_pJET_RFP_DsRed_iGEM_Goettingen_2015.jpg" style="; float:right;"> | <img src=" https://static.igem.org/mediawiki/2015/1/1d/Restr_pJET_RFP_DsRed_iGEM_Goettingen_2015.jpg" style="; float:right;"> | ||
| − | |||
| − | |||
<p> </p> | <p> </p> | ||
<p><strong>RESULT</strong></p> | <p><strong>RESULT</strong></p> | ||
| Line 269: | Line 268: | ||
</div> | </div> | ||
| − | + | ||
Revision as of 07:21, 16 September 2015
Project Results
Transformation Efficiency Kit, RFP construct (iGEM)
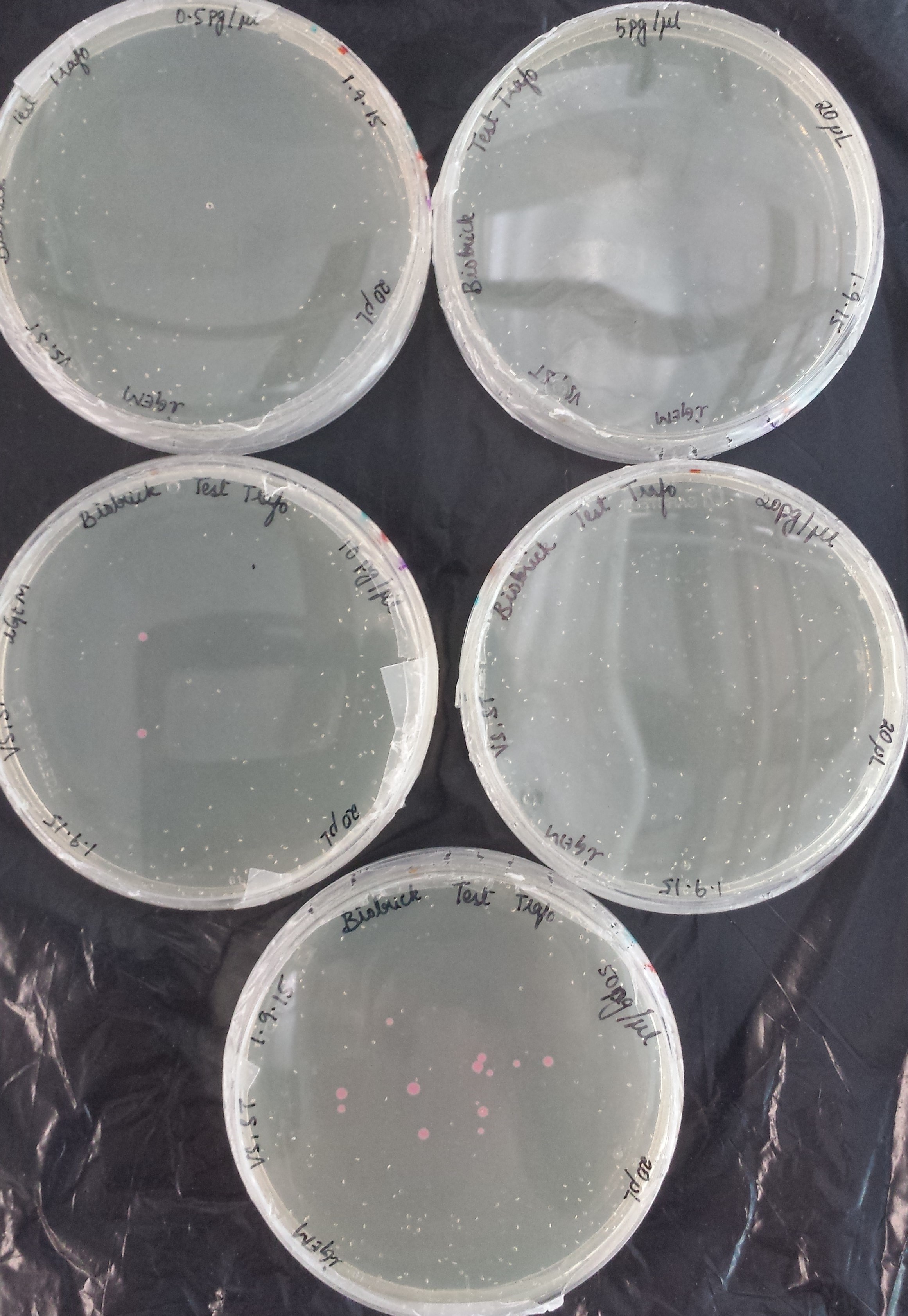
Before using our competent E. coli TOP10 cells in the important experiments, we used the Transformation Efficiency Kit to test the efficiency of our competent cells!
The kit includes five vials of each different DNA concentration: 50pg/μl, 20pg/μl, 10pg/μl, 5pg/μl, 0.5pg/μl of purified DNA from BBa_J04450 (RFP construct) in plasmid backbone pSB1C3.
The first test transformation (50 μl of competent cells, 20 μl plated) showed very poor results:
|
DNA concentration |
0.5pg/μl |
5pg/μl |
10pg/μl |
20pg/μl |
50pg/μl |
|
# of colonies |
0 |
0 |
2 |
0 |
13 |
|
efficiency (cfu) |
0 |
0 |
3.8x106 |
0 |
3.4x106 |
|
Results |
efficiency |
|
average |
1.4x106 |
|
weighted |
3.5x106 |
The second test transformation (200 μl of competent cells, 100 μl plated) showed still poor results but we decided to continue working with our cells:

|
DNA concentration |
0.5pg/μl |
5pg/μl |
10pg/μl |
20pg/μl |
50pg/μl |
|
# of colonies |
1 |
13 |
1 |
35 |
28 |
|
efficiency (cfu) |
8.0x106 |
1.0x107 |
4.0x105 |
7.0x106 |
2.3x106 |
|
Results |
efficiency |
|
average |
5.6x106 |
|
weighted |
3.7x106 |
RFP construct
RFP construct
RFP (RFP DsRed) was amplified from by PCR (Fig.1). Colonies on a plate were given to us by the Applied and Genomic Microbiology department. Primers contained restriction sites for KpnI and SacI in order to make them compatible for insertion into the multiple cloning site of the pBAD A vector.

After purification of the PCR product it was ligated into pJET1.2 by subcloning (blunt end ligation). This vector serves to clone the fluorescent protein without triggering its activity, which may interact with the expression vector or the chosen E.coli strain.
Ligation into pJET1.2 was followed according to the protocol in the methods collection. After over-night incubation colonies were picked, plasmids extracted with the QIAGEN QIAprep Spin Miniprep Kit and restricted with KpnI and SacI.
Once restriction controls showed the correct bands, both pJET_RFP_3 and pJET_RFP_7 were sent for sequencing by the G2L laboratory.

RESULT
Both sequences were correct. We decided to work with pJET_RFP_3. The plasmid was transformed with E.coli TOP10 and E.coli BL21. Cryocultures were frozen for a strain collection.
Afterwards the RFP_3 insert was ligated into pBAD A via the T4 ligase system according to the protocol in the methods collection and transformed into E.coli TOP10.
MICROSCOPY
To check the red fluorescence transformed E.coli TOP10 were examined by fluorescence microscopy with an excitation by 536 nm. RFP DsRed should show an emission by 582 nm.
Unfortunately no fluorescence could be detected throughout the whole project. Nevertheless the DNA sequence of the construct is correct. It might be a problem of expression.
Here you can describe the results of your project and your future plans.
What should this page contain?
- Clearly and objectively describe the results of your work.
- Future plans for the project
- Considerations for replicating the experiments
Project Achievements
You can also include a list of bullet points (and links) of the successes and failures you have had over your summer. It is a quick reference page for the judges to see what you achieved during your summer.
- A list of linked bullet points of the successful results during your project
- A list of linked bullet points of the unsuccessful results during your project. This is about being scientifically honest. If you worked on an area for a long time with no success, tell us so we know where you put your effort.
Inspiration
See how other teams presented their results.