Difference between revisions of "Team:Goettingen/Experiments"
| Line 466: | Line 466: | ||
<p>Cloning principle:</p> | <p>Cloning principle:</p> | ||
<ul> | <ul> | ||
| − | <li>pJET1.2 is a linearized cloning vector | + | <li>pJET1.2 is a linearized cloning vector designed for inserts from 6 bp to 10 kb. The 5’-ends of the vector contain phosporyl groups, therefore phosphorylation of the PCR products is indicated.</li> |
</ul> | </ul> | ||
<ul> | <ul> | ||
| Line 472: | Line 472: | ||
</ul> | </ul> | ||
<ul> | <ul> | ||
| − | <li>Optimal PCR product quantity for ligation reaction is to be calculated from the Kit protocol, For the length of | + | <li>Optimal PCR product quantity for ligation reaction is to be calculated from the Kit protocol, For the length of 2.5 kb of PCR product, to have 0.15 pmol ends of the PCR product in the ligation reaction 125 ng of the PCR product should be used.</li> |
</ul> | </ul> | ||
<ul> | <ul> | ||
| Line 535: | Line 535: | ||
<ul> | <ul> | ||
| − | <li>You will perform TOPO® Cloning in a reaction buffer containing salt. Note | + | <li>You will perform TOPO® Cloning in a reaction buffer containing salt. Note: the amount of salt added to the TOPO® Cloning reaction has to be adapted to the planned transformation protocol (chemically competent cells or electrocompetent cells).</li> |
</ul> | </ul> | ||
<p><strong> </strong>- Set up the TOPO® Cloning reaction depending on the transformation method.</p> | <p><strong> </strong>- Set up the TOPO® Cloning reaction depending on the transformation method.</p> | ||
| Line 586: | Line 586: | ||
<tr> | <tr> | ||
<td valign="top" width="174"> | <td valign="top" width="174"> | ||
| − | <p>Sterile | + | <p>Sterile water</p> |
</td> | </td> | ||
<td valign="top" width="212"> | <td valign="top" width="212"> | ||
| Line 655: | Line 655: | ||
<p>- Transfer the culture without any air bubbles into pre-cooled electroporation cuvettes (40 µl maximum) and incubate on ice for 10 minutes.</p> | <p>- Transfer the culture without any air bubbles into pre-cooled electroporation cuvettes (40 µl maximum) and incubate on ice for 10 minutes.</p> | ||
<p>- Electroporate using the electroporator with 1.25 mV, 5 decharge time.</p> | <p>- Electroporate using the electroporator with 1.25 mV, 5 decharge time.</p> | ||
| − | <p>- Immediately after electroporation, transfer 300 µl room temperature LB medium on top of the cells and transfer it into an 1. | + | <p>- Immediately after electroporation, transfer 300 µl room temperature LB medium on top of the cells and transfer it into an 1.5 ml fresh E-cup</p> |
<p>- Incubate the culture for 1 hour at 37°C and 150 rpm</p> | <p>- Incubate the culture for 1 hour at 37°C and 150 rpm</p> | ||
<p><sup>- </sup>Spread a 100 µl from the dilution series (10<sup>-3</sup> to 10<sup>-6</sup>) on a pre-warmed LB plate containing ampicillin</p> | <p><sup>- </sup>Spread a 100 µl from the dilution series (10<sup>-3</sup> to 10<sup>-6</sup>) on a pre-warmed LB plate containing ampicillin</p> | ||
| Line 669: | Line 669: | ||
<li>Add the provided RNase A solution to buffer P1, mix, and store at 4 <sup>o</sup>C.</li> | <li>Add the provided RNase A solution to buffer P1, mix, and store at 4 <sup>o</sup>C.</li> | ||
<li>Add ethanol (96–100%) to Buffer PE before use.</li> | <li>Add ethanol (96–100%) to Buffer PE before use.</li> | ||
| − | <li>All centrifugation steps are carried out at | + | <li>All centrifugation steps are carried out at 17,900 x g (13,000 rpm) in a conventional table-top microcentrifuge at room temperature.</li> |
</ul> | </ul> | ||
<p> </p> | <p> </p> | ||
| Line 676: | Line 676: | ||
<p>- Inoculate the medium with the desired <em>E.coli</em> strain and incubate overnight at 37 <sup>o</sup>C with agitation (150 rpm).</p> | <p>- Inoculate the medium with the desired <em>E.coli</em> strain and incubate overnight at 37 <sup>o</sup>C with agitation (150 rpm).</p> | ||
| − | <p>- Pellet the overnight culture by centrifugation at | + | <p>- Pellet the overnight culture by centrifugation at 8,000 rpm (6,800xg) for 3 min at room temperature.</p> |
<p>- Resuspend pelleted bacterial cells in 250 μl Buffer P1 and transfer it to a microcentrifuge tube.</p> | <p>- Resuspend pelleted bacterial cells in 250 μl Buffer P1 and transfer it to a microcentrifuge tube.</p> | ||
| Line 684: | Line 684: | ||
<p>- Add 350 μl Buffer N3 and mix immediately and thoroughly by inverting the tube 4–6 times.</p> | <p>- Add 350 μl Buffer N3 and mix immediately and thoroughly by inverting the tube 4–6 times.</p> | ||
| − | <p>- Centrifuge for 10 min at | + | <p>- Centrifuge for 10 min at 17,900 x g in a table-top microcentrifuge.</p> |
<p>- Apply the supernatant to the QIAprep spin column by decanting. Centrifuge 60 s. Discard the flow-through.</p> | <p>- Apply the supernatant to the QIAprep spin column by decanting. Centrifuge 60 s. Discard the flow-through.</p> | ||
| Line 716: | Line 716: | ||
</p> | </p> | ||
<p> | <p> | ||
| − | Centrifuge the culture at | + | Centrifuge the culture at 10,000 x g for 2 min to obtain the pellet and repeat the process until the culture is completely centrifuged. Store the pellet of |
1 ml of the culture at -20°C for future use. | 1 ml of the culture at -20°C for future use. | ||
</p> | </p> | ||
| Line 728: | Line 728: | ||
<p> | <p> | ||
Add 350 µl of Solution III to the cleared lysate and gently mix by inverting the tubes 6 -10 times until a flocculent white precipitate is formed. | Add 350 µl of Solution III to the cleared lysate and gently mix by inverting the tubes 6 -10 times until a flocculent white precipitate is formed. | ||
| − | Centrifuge at | + | Centrifuge at 10,000 x g for 10 min at room temperature. |
</p> | </p> | ||
<p> | <p> | ||
Transfer the clear supernatant to a fresh PerfectBind DNA Column in a 2 ml Collection Tube. Centrifuge the Column with the Collection Tube for 1 min at | Transfer the clear supernatant to a fresh PerfectBind DNA Column in a 2 ml Collection Tube. Centrifuge the Column with the Collection Tube for 1 min at | ||
| − | + | 10,000 x g at room temperature. Discard the flow-through liquid. | |
</p> | </p> | ||
<p> | <p> | ||
| − | Add 500 µl of PW Plasmid buffer to the PerfectBind DNA Column in the Collection Tube and centrifuge for 1 min at | + | Add 500 µl of PW Plasmid buffer to the PerfectBind DNA Column in the Collection Tube and centrifuge for 1 min at 10,000 x g. Discard the flow-through. |
</p> | </p> | ||
<p> | <p> | ||
| − | Add 750 µl of DNA Wash buffer to the PerfectBind DNA Column in the Collection tube and centrifuge for 1 min at | + | Add 750 µl of DNA Wash buffer to the PerfectBind DNA Column in the Collection tube and centrifuge for 1 min at 10,000 x g. Discard the flow-through. Repeat |
this step to obtain optimum results. | this step to obtain optimum results. | ||
</p> | </p> | ||
<p> | <p> | ||
| − | Place the PerfectBind DNA Column in the Collection tube and centrifuge for 2 min at | + | Place the PerfectBind DNA Column in the Collection tube and centrifuge for 2 min at 10,000 x g to dry the column matrix. This step is essential to remove |
ethanol from the column. | ethanol from the column. | ||
</p> | </p> | ||
<p> | <p> | ||
Place the PerfectBind DNA Column into a fresh 1.5 ml Eppendorf tube. Add 50 µl of pre-warmed sterile deionized water directly to the binding matrix in the | Place the PerfectBind DNA Column into a fresh 1.5 ml Eppendorf tube. Add 50 µl of pre-warmed sterile deionized water directly to the binding matrix in the | ||
| − | PerfectBind DNA Column and centrifuge for 1 min at | + | PerfectBind DNA Column and centrifuge for 1 min at 5,000 x g to elute the DNA. |
</p> | </p> | ||
<p> | <p> | ||
| Line 769: | Line 769: | ||
<p> pH 7.5 with acetic acid</p> | <p> pH 7.5 with acetic acid</p> | ||
<p>Buffer RF2 (80 ml): 1.6 ml MOPS (0.5 M) stock solution</p> | <p>Buffer RF2 (80 ml): 1.6 ml MOPS (0.5 M) stock solution</p> | ||
| − | <p> | + | <p> pH 6.8 with NaOH</p> |
<p> 0.096 g RbCl</p> | <p> 0.096 g RbCl</p> | ||
<p> 0.88 g CaCl<sub>2</sub>.2H<sub>2</sub>O</p> | <p> 0.88 g CaCl<sub>2</sub>.2H<sub>2</sub>O</p> | ||
| Line 775: | Line 775: | ||
<p>- Inoculate a 4 ml culture either with a single colony or with a cryoculture of the desired <em>E.coli </em>strainand incubate the culture with agitation overnight at 37°C</p> | <p>- Inoculate a 4 ml culture either with a single colony or with a cryoculture of the desired <em>E.coli </em>strainand incubate the culture with agitation overnight at 37°C</p> | ||
<p><sub>- </sub>Inoculate a 300 ml shake flask containing 100 ml LB medium with the overnight culture to an OD<sub>600</sub> of 0.05 and grow the culture at 37°C until the OD<sub>600</sub>is about 0.3</p> | <p><sub>- </sub>Inoculate a 300 ml shake flask containing 100 ml LB medium with the overnight culture to an OD<sub>600</sub> of 0.05 and grow the culture at 37°C until the OD<sub>600</sub>is about 0.3</p> | ||
| − | <p>- Transfer the cells into two 50 ml falcon tubes, incubate the cultures for 15 minues on ice and harvest the cells by centrifugation for 15 min at | + | <p>- Transfer the cells into two 50 ml falcon tubes, incubate the cultures for 15 minues on ice and harvest the cells by centrifugation for 15 min at 5,000 rpm and 4°C. Discard the supernatants.</p> |
| − | <p>- Resuspend the cells in 1/3 of the original volume (~16 ml/50 ml) of buffer RF1, incubate the cells again on ice and harvest the cells by centrifugation for 15 min at | + | <p>- Resuspend the cells in 1/3 of the original volume (~16 ml/50 ml) of buffer RF1, incubate the cells again on ice and harvest the cells by centrifugation for 15 min at 5,000 rpm and 4°C. Discard the supernatants.</p> |
<p>- Resuspend the cells in 4 ml of buffer RF2 and incubate the suspensions for 15 min on ice. Prepare the Eppendorf tubes and liquid nitrogen.</p> | <p>- Resuspend the cells in 4 ml of buffer RF2 and incubate the suspensions for 15 min on ice. Prepare the Eppendorf tubes and liquid nitrogen.</p> | ||
<p>- Put 0.4 ml of the cell suspension into the Eppendorf reaction tubes and freeze the cells by transferring them immediately to the liquid nitrogen. Store the competent cells at -80°C</p> | <p>- Put 0.4 ml of the cell suspension into the Eppendorf reaction tubes and freeze the cells by transferring them immediately to the liquid nitrogen. Store the competent cells at -80°C</p> | ||
| Line 802: | Line 802: | ||
<p>- Inoculate a 20 ml culture either with a single colony or with the cryoculture of the desired <em>E.coli </em>strainand incubate the culture with agitation for 20 h at 28°C</p> | <p>- Inoculate a 20 ml culture either with a single colony or with the cryoculture of the desired <em>E.coli </em>strainand incubate the culture with agitation for 20 h at 28°C</p> | ||
<p>- Inoculate a 250 ml SOB medium supplemented in a 2 l shake flask and grow the cells to an OD<sub>600</sub> of 0.5- 0.9 (20-20 h) at 18°C and 200-250 rpm</p> | <p>- Inoculate a 250 ml SOB medium supplemented in a 2 l shake flask and grow the cells to an OD<sub>600</sub> of 0.5- 0.9 (20-20 h) at 18°C and 200-250 rpm</p> | ||
| − | <p>- Incubate the whole flask for 10 min on ice. Collect the cells by centrifugation for 10 min at 4°C and 5000 rpm. Resuspend the cells in 80 ml of ice-cold TB and incubate them for 10 min on ice. Collect the cells by centrifugation for 5 min at | + | <p>- Incubate the whole flask for 10 min on ice. Collect the cells by centrifugation for 10 min at 4°C and 5000 rpm. Resuspend the cells in 80 ml of ice-cold TB and incubate them for 10 min on ice. Collect the cells by centrifugation for 5 min at 5,000 rpm.</p> |
<p>- Resuspend the cells in a 20 ml of ice-cold TB. Add DMSO to a final concentration of 7% (1.4 ml) and gently shake the falcon tube.</p> | <p>- Resuspend the cells in a 20 ml of ice-cold TB. Add DMSO to a final concentration of 7% (1.4 ml) and gently shake the falcon tube.</p> | ||
<p>- Transfer 0.2 ml aliquots into labelled eppendorf reaction tubes and freeze the cells in liquid nitrogen. Store the cells at –80°C.</p> | <p>- Transfer 0.2 ml aliquots into labelled eppendorf reaction tubes and freeze the cells in liquid nitrogen. Store the cells at –80°C.</p> | ||
| Line 824: | Line 824: | ||
<p>Pipet 20 µL from each tube onto the appropriate plate, and spread the mixture evenly across the plate. Do triplicates (3 each) of each tube if possible, so you can calculate an average colony yield. Incubate at 37°C overnight. Position the plates so the agar side is facing up, and the lid is facing down.</p> | <p>Pipet 20 µL from each tube onto the appropriate plate, and spread the mixture evenly across the plate. Do triplicates (3 each) of each tube if possible, so you can calculate an average colony yield. Incubate at 37°C overnight. Position the plates so the agar side is facing up, and the lid is facing down.</p> | ||
<p>Count the number of colonies on a light field or a dark background, such as a lab bench. Use the following equation to calculate your competent cell efficiency. If you've done triplicates of each sample, use the average cell colony count in the calculation.</p> | <p>Count the number of colonies on a light field or a dark background, such as a lab bench. Use the following equation to calculate your competent cell efficiency. If you've done triplicates of each sample, use the average cell colony count in the calculation.</p> | ||
| − | <p>(colonies on plate) / ng of DNA plated x | + | <p>(colonies on plate) / ng of DNA plated x 1,000ng/µg</p> |
<p>Note: The measurement "ng of DNA plated" refers to how much DNA was plated onto each agar plate, not the total amount of DNA used per transformation. You can calculate this number using the following equation:</p> | <p>Note: The measurement "ng of DNA plated" refers to how much DNA was plated onto each agar plate, not the total amount of DNA used per transformation. You can calculate this number using the following equation:</p> | ||
<p>1 µL x concentration of DNA (refer to vial) x (volume plated / total reaction volume)</p> | <p>1 µL x concentration of DNA (refer to vial) x (volume plated / total reaction volume)</p> | ||
| Line 847: | Line 847: | ||
</td> | </td> | ||
<td valign="top" width="97"> | <td valign="top" width="97"> | ||
| − | <p>7. | + | <p>7.8 g</p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 855: | Line 855: | ||
</td> | </td> | ||
<td valign="top" width="97"> | <td valign="top" width="97"> | ||
| − | <p>17. | + | <p>17.5 g</p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 863: | Line 863: | ||
</td> | </td> | ||
<td valign="top" width="97"> | <td valign="top" width="97"> | ||
| − | <p>ad. | + | <p>ad. 1000 mL</p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 878: | Line 878: | ||
</td> | </td> | ||
<td valign="top" width="97"> | <td valign="top" width="97"> | ||
| − | <p>7. | + | <p>7.8 g</p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 886: | Line 886: | ||
</td> | </td> | ||
<td valign="top" width="97"> | <td valign="top" width="97"> | ||
| − | <p>17. | + | <p>17.5 g</p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 894: | Line 894: | ||
</td> | </td> | ||
<td valign="top" width="97"> | <td valign="top" width="97"> | ||
| − | <p>17. | + | <p>17.0 g</p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 902: | Line 902: | ||
</td> | </td> | ||
<td valign="top" width="97"> | <td valign="top" width="97"> | ||
| − | <p>ad. | + | <p>ad. 1,000 mL</p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 911: | Line 911: | ||
<p> </p> | <p> </p> | ||
<p><strong>1.) Cell extraction by French Press:</strong></p> | <p><strong>1.) Cell extraction by French Press:</strong></p> | ||
| − | <p>- Resuspend pellet in | + | <p>- Resuspend pellet in 20 mL 1x LEW Buffer from the kit</p> |
| − | <p>- Take a small sample (10µl) for microscopy</p> | + | <p>- Take a small sample (10 µl) for microscopy</p> |
<p>- Press 4 times with a French Press at High Pressure and collect the flow through</p> | <p>- Press 4 times with a French Press at High Pressure and collect the flow through</p> | ||
<p>- Take a small sample (10µl) and analyse both before and after press samples under the microscope. Look for inclusion bodies.</p> | <p>- Take a small sample (10µl) and analyse both before and after press samples under the microscope. Look for inclusion bodies.</p> | ||
| − | <p>- Centrifuge pressed samples at 4°C and | + | <p>- Centrifuge pressed samples at 4°C and 13,000 rpm for 45 minutes, keep supernatant</p> |
<p> </p> | <p> </p> | ||
| − | <p><strong>2.) Protino® Ni-IDA | + | <p><strong>2.) Protino® Ni-IDA 2,000 His-Tag protein purification (Macherey-Nagel)</strong></p> |
| − | <p>- Wet Ni-IDA column with | + | <p>- Wet Ni-IDA column with 4 mL of 1x LEW buffer and discard flow through</p> |
<p>- Run supernatant from the cell extraction through the column and collect flow through (cell extract, store at 4°C)</p> | <p>- Run supernatant from the cell extraction through the column and collect flow through (cell extract, store at 4°C)</p> | ||
| − | <p>- Wash column 3 times with | + | <p>- Wash column 3 times with 4 mL 1x LEW Buffer</p> |
<p>- Collect each flow through (wash 1-3) and store at 4°C</p> | <p>- Collect each flow through (wash 1-3) and store at 4°C</p> | ||
| − | <p>- Elute protein 3 times with | + | <p>- Elute protein 3 times with 3 mL Elution Buffer (contains Imidazole)</p> |
<p>- Collect each elution and store fractions at 4°C</p> | <p>- Collect each elution and store fractions at 4°C</p> | ||
<p>- Quantify protein content by Bradford measurement (see Bradford assay)</p> | <p>- Quantify protein content by Bradford measurement (see Bradford assay)</p> | ||
| Line 1,018: | Line 1,018: | ||
<div id="menu9"> | <div id="menu9"> | ||
<p>Materials:</p> | <p>Materials:</p> | ||
| − | <p>Substrate- 1mM 4-nitrophenyl butyrate prepared in Na-Phosphate buffer ( | + | <p>Substrate- 1mM 4-nitrophenyl butyrate prepared in Na-Phosphate buffer (pH 8.0)</p> |
<p>Protein sample</p> | <p>Protein sample</p> | ||
<p>Procedure:</p> | <p>Procedure:</p> | ||
| Line 1,146: | Line 1,146: | ||
</td> | </td> | ||
<td valign="top"> | <td valign="top"> | ||
| − | <p>2µl </p> | + | <p>2 µl </p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 1,154: | Line 1,154: | ||
</td> | </td> | ||
<td valign="top"> | <td valign="top"> | ||
| − | <p>1µl </p> | + | <p>1 µl </p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 1,190: | Line 1,190: | ||
<li>For 10 µl reaction add 1 µl of 10x Tango buffer, for 20 µl reaction add 2 µl of 10x Tango buffer and vice versa.</li> | <li>For 10 µl reaction add 1 µl of 10x Tango buffer, for 20 µl reaction add 2 µl of 10x Tango buffer and vice versa.</li> | ||
<li>Incubate at 37 <sup>o</sup>C for 2 hours.</li> | <li>Incubate at 37 <sup>o</sup>C for 2 hours.</li> | ||
| − | <li>For inactivation incubation at 80°C for | + | <li>For inactivation incubation at 80°C for 5 min. </li> |
<li>Check the products on 0.8% agarose gel.</li> | <li>Check the products on 0.8% agarose gel.</li> | ||
</ul> | </ul> | ||
| Line 1,211: | Line 1,211: | ||
</td> | </td> | ||
<td valign="top" width="96"> | <td valign="top" width="96"> | ||
| − | <p>500- | + | <p>500-1,000 ng</p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 1,270: | Line 1,270: | ||
</td> | </td> | ||
<td valign="top"> | <td valign="top"> | ||
| − | <p>500- | + | <p>500-1,000 ng</p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 1,424: | Line 1,424: | ||
</td> | </td> | ||
<td valign="top" width="301"> | <td valign="top" width="301"> | ||
| − | <p> | + | <p>1,000 ng (2 µl)</p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 1,432: | Line 1,432: | ||
</td> | </td> | ||
<td valign="top" width="301"> | <td valign="top" width="301"> | ||
| − | <p>1µl</p> | + | <p>1 µl</p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 1,464: | Line 1,464: | ||
</td> | </td> | ||
<td valign="top"> | <td valign="top"> | ||
| − | <p>15µl</p> | + | <p>15 µl</p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 1,472: | Line 1,472: | ||
</td> | </td> | ||
<td valign="top"> | <td valign="top"> | ||
| − | <p>2µl</p> | + | <p>2 µl</p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 1,480: | Line 1,480: | ||
</td> | </td> | ||
<td valign="top"> | <td valign="top"> | ||
| − | <p>2µl</p> | + | <p>2 µl</p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 1,488: | Line 1,488: | ||
</td> | </td> | ||
<td valign="top"> | <td valign="top"> | ||
| − | <p>1µl</p> | + | <p>1 µl</p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 1,496: | Line 1,496: | ||
</td> | </td> | ||
<td valign="top"> | <td valign="top"> | ||
| − | <p>1µl</p> | + | <p>1 µl</p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 1,502: | Line 1,502: | ||
</table> | </table> | ||
<p> </p> | <p> </p> | ||
| − | <p>Incubate | + | <p>Incubate 20 min at 37˚C and inactivate 5 min at 80˚C.</p> |
<p> </p> | <p> </p> | ||
<p></p> | <p></p> | ||
| Line 1,595: | Line 1,595: | ||
</td> | </td> | ||
<td valign="top" width="57"> | <td valign="top" width="57"> | ||
| − | <p> | + | <p>12 ul</p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 1,603: | Line 1,603: | ||
</td> | </td> | ||
<td valign="top" width="57"> | <td valign="top" width="57"> | ||
| − | <p> | + | <p>2 ul</p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 1,611: | Line 1,611: | ||
</td> | </td> | ||
<td valign="top" width="57"> | <td valign="top" width="57"> | ||
| − | <p> | + | <p>2 ul</p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 1,627: | Line 1,627: | ||
</td> | </td> | ||
<td valign="top" width="57"> | <td valign="top" width="57"> | ||
| − | <p> | + | <p>1 ul</p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 1,635: | Line 1,635: | ||
</td> | </td> | ||
<td valign="top" width="57"> | <td valign="top" width="57"> | ||
| − | <p> | + | <p>1 ul</p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 1,671: | Line 1,671: | ||
<ul> | <ul> | ||
<p>This protocol describes PCR procedure using Phusion High-Fidelity DNA Polymerase. <strong>(Thermo Fisher Scientific).</strong></p> | <p>This protocol describes PCR procedure using Phusion High-Fidelity DNA Polymerase. <strong>(Thermo Fisher Scientific).</strong></p> | ||
| − | <p>• | + | <p>• Note: the annealing conditions depend on the applied DNA polymerases (such as Taq DNA polymerases).</p> |
<p>•Use 15–30 s/kb for extension. Do not exceed 1 min/kb.</p> | <p>•Use 15–30 s/kb for extension. Do not exceed 1 min/kb.</p> | ||
<p>•Phusion DNA Polymerases produce blunt end DNA products.</p> | <p>•Phusion DNA Polymerases produce blunt end DNA products.</p> | ||
| Line 1,928: | Line 1,928: | ||
</td> | </td> | ||
<td valign="top" nowrap="nowrap" width="96"> | <td valign="top" nowrap="nowrap" width="96"> | ||
| − | <p align="center"> | + | <p align="center">10 s</p> |
</td> | </td> | ||
<td rowspan="3" valign="top" nowrap="nowrap" width="88"> | <td rowspan="3" valign="top" nowrap="nowrap" width="88"> | ||
| Line 1,942: | Line 1,942: | ||
</td> | </td> | ||
<td valign="top" nowrap="nowrap" width="96"> | <td valign="top" nowrap="nowrap" width="96"> | ||
| − | <p align="center"> | + | <p align="center">15 s</p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 2,158: | Line 2,158: | ||
<ul> | <ul> | ||
<p><strong>RFP microscopy</strong></p> | <p><strong>RFP microscopy</strong></p> | ||
| − | <p>To check if the transformed <em>E.coli</em> TOP10 show red fluorescence, the culture was examined by fluorescence microscopy. The RFP DsRed filter was used (excitation at 536 nm, emission at 582 nm). To prevent the cells from floating around they are fixed in 0.8 %.</p> | + | <p>To check if the transformed <em>E.coli</em> TOP10 show red fluorescence, the culture was examined by fluorescence microscopy. The RFP DsRed filter was used (excitation at 536 nm, emission at 582 nm). To prevent the cells from floating around they are fixed in 0.8%.</p> |
<p> </p> | <p> </p> | ||
<p><strong>Preparation of slides</strong></p> | <p><strong>Preparation of slides</strong></p> | ||
<ul> | <ul> | ||
| − | <li>Prepare 0.8 % agarose with water and boil it up.</li> | + | <li>Prepare 0.8% agarose with water and boil it up.</li> |
<li>Pipet 500 μL onto a slide and press another one on top. Let the agarose cure between both slides.</li> | <li>Pipet 500 μL onto a slide and press another one on top. Let the agarose cure between both slides.</li> | ||
<li>Carefully remove the upper slide and put the sample name in one corner the one with the agarose.</li> | <li>Carefully remove the upper slide and put the sample name in one corner the one with the agarose.</li> | ||
Revision as of 13:46, 18 September 2015
Media/Buffer
LB Medium
"Fat" LB Medium
Phosphatase Activity plates, Sperber media
Esterase Activity plates, with 1% Tributyrin
Cellulase activity plates
1x TAE Buffer
Cloning Methods
PCR product purification using QIAquick® PCR Purification Kit (QIAGEN)
PCR Gel extraction, peqGOLD Gel Extraction Kit
Blunt End Ligation in pJET1.2 vector –Clone JET PCR Cloning Kit– (Thermo Scientific)
Sticky End T4 Ligation (Thermo Scientific)
TOPO® Cloning protocol usingChampion™ pET Directional TOPO® Expression Kits (Thermo Fisher Scientific)
Plasmid transformation into chemically competent E. coli
Electroporation of BL21 cells with pJET_RFP
Plasmid Extraction - using QIAprep Spin Miniprep Kit (QIAGEN)
Plasmid Extraction - using peqGOLD Plasmid Miniprep Kit I (PEQLAB Technologies)
Competent Cells
Preparation of competent E.coli cells
Transformation Efficiency Kit, RFP construct (iGEM)
Protein Extraction and Purification
Protein Extraction (French Press) and Purification (Protino® Ni-IDA 2000 His-Tag protein purification, Macherey-Nagel)
Affinity chromatography of His-tagged proteins
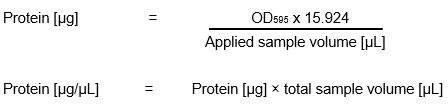
Bradford Assay
Activity Screens
Esterase activity test
Phosphatase activity test
Cellulase activity screening
Restriction Controls
Aan I (Psi I ) - thermo fisher scientific - restriction control protocol
Double digestion restriction control
Restriction control using fast and slow digestion enzymes
Scafoldin Restriction control
Esterase Restriction Control
Phosphatase Restriction Control
PCR Preparation Methods
Colony PCR
Phusion PCR
Sequencing
Protocol for Sanger sequencing
Overnight Sanger Sequencing
Fluorescence Microscopy
RFP microscopy