Difference between revisions of "Team:ETH Zurich/Collaborations"
| Line 30: | Line 30: | ||
</html> | </html> | ||
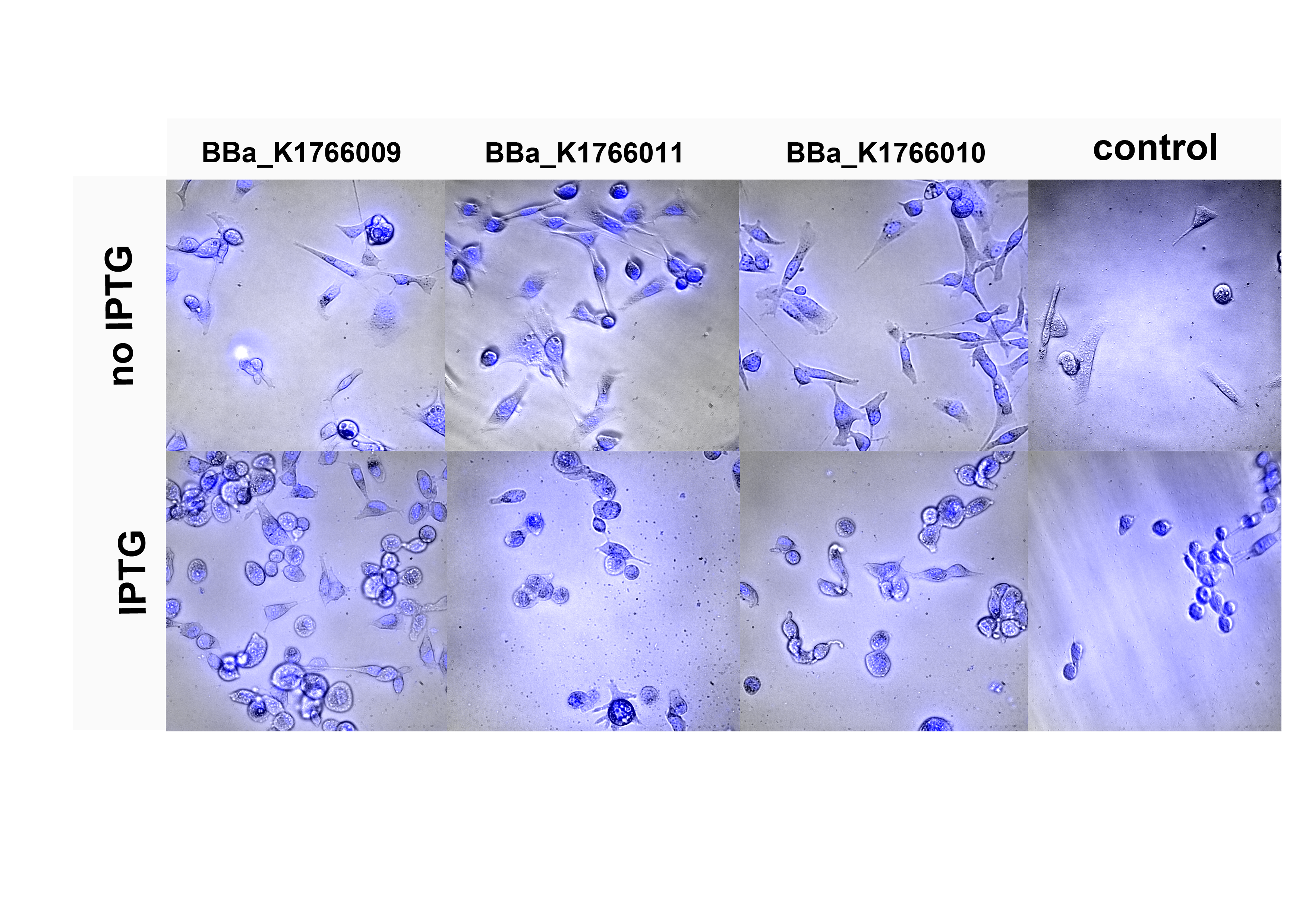
| − | [[File:ETH15_stockholm_collaboration2.png|thumb|600px|center|Figure 1 | + | [[File:ETH15_stockholm_collaboration2.png|thumb|600px|center|<b>Figure 1.</b> We tested three different constructs for their binding to HER2-positive SK-BR-3 cells. The comparison of induced and non-induced spheroblasts did not yeald conclusive results.]] |
<html> | <html> | ||
Revision as of 00:24, 19 September 2015
- Project
- Modeling
- Lab
- Human
Practices - Parts
- About Us
Collaborations
One of the biggest opportunities in iGEM is the contact and collaboration between teams of different backgrounds, which makes it possible to get input from various points of views and to establish a network of young scientist with the same interests. We took advantage of this opportunity and established contacts to teams from several continents.
Stockholm
Starting from the very beginning of our iGEM summer we were in continuous contact with the team from Stockholm. We realized soon that our projects were going into a similar direction, both focusing on cancer detection in samples of bodily fluids, using engineered bacteria that will bind to cancer cells. In contrast to our project, Stockholm’s ABBBA is focussed on the detection of one specific type of cancer at a time. In several Skype meetings we exchanged our ideas about the best possible setup for single cell analysis and signal integration of the bacteria upon binding to a cancer cell. We provided the team from Stockholm with inputs on microfluidics as a setup for the final analysis and they helped us when we were struggling with the recovery of our gene fragments. But what seemed to be the best opportunity for combining our forces was the rather extensive experience in mammalian cell culture and bacterial-mammalian coculture our team had gathered in the course of our project. Since the team of Stockholm did not have the opportunity to test their system in such a real-life system, we decided to validate their system by combining their engineered E. coli cells with their actual targets: HER2-positive breast cancer cells.
Validation of ABBBA binding to cancer cells
We characterized the system of Stockholm regarding the binding of their bacteria to HER2-positive breast cancer cells. The affibody based system explored by the Stockholm iGEM team is designed to make engineered E. coli cells bind to cancer cells expressing the targeted surface marker. In the tested case of HER2-positive breast cancer, the team came up with three chimeric receptors, targeting different epitopes of the HER2 extracellular domain. We tested the binding of ABBBA E. coli expressing the respective constructs to HER2+ SK-BR-3 cells.
For better observation of colocalization of the bacteria and the mammalian cells we tried to introduce a second plasmid carrying a constitutively expressed fluorophore into the bacterial strains that we received from Stockholm. Unfortunately, all atempts to this failed. As an alternative, we tried transforming the plasimds carrying their test constructs into E. coli expressing GFP from their genome. It seems that cells are not quite happy accepting the plasmids of interest, which was surprising since we never had probems with any of our transformations. Nevertheless, we were determined to give our best in this cause and went on to characterize the interaction of bacteria and mammalian cells after aplying a spheroblast protocol and inducing the bacteria with IPTG for expression of the LacI-regulated constructs by microscopy. The spheroblast protocol is necessary since the affibody construct is localized in the inner membrane of E. coli and has to be revealed by removal of the outer membrane.
Results
Unfortunately, the lack of a way to clearly identify the ABBBA bacteria via a marker made it very hard to differentiate between bound bacteria and bumps in the surface of mammalian cells. We tried to visualize an interaction but we have to say that with the methods available to us all our atempts could not give us conclusive results.
Characterization of lldR-operator promoter collection
In exchange, Stockholm agreed to characterize a part of our synthetically designed LldR-operator promoter collection for us. They probed our engineered bacteria, containing a green fluorescent protein under the control of lldR along with constantly expressed lldR with or without lldP, for their reaction to different levels of L-Lactate, thereby greatly contributing to the characterization of our promoter collection.
Amoy
We collaborated with the Amoy team by contributing to their newsletter. We have written about the selection process for iGEM in the issue No1 and participated in the questionnaire about the most useful software for us in issue No6. We also published a description of our project in issue No3, an update in issue No5 and about our human practices in issue No7. Finally, we wrote short essays about the situation of synthetic biology in Switzerland for the Special issue and about the ethical dilemmas of editing a human embryo for the issue No2. Next to the Amoy team itself, we were the only participating team that contributed to every issue of the newsletter.
Darmstadt
On their southern Germany tour, representatives of the TU_Darmstadt iGEM team visited several German, Swiss, and Italian teams. We were happy to host them for a weekend in Zurich and Basel, where we showed them our lab and our city. We were talking a lot about our projects which was a great opportunity to find out how someone completely unrelated to our idea would react to it, and vice versa.
Next to this, TU_Darmstadt also introduced us to their Lab Surfing platform, which our team tested for them and was able to come up with a lot of inputs to improve the user interface for this great idea. The result of the TU_Darmstadt Lab Surfing project can be found .

EPFL
Since both Swiss Federal Institutes of Technology, ETH Zurich and EPF Lausanne, are hosting an iGEM team in 2015, we were of course eager to find out what our colleagues in Lausanne were working on. We invited their team to our lab where we were able to test our presentations on each other and get useful inputs on how to improve them.
We found also that for both our teams it is important to spread public awareness on the rapidly rising field of synthetic biology to which we have become so used. EPFL invited us to take part in their survey on the public awareness of and opinion on synthetic biology, which they had planned to conduct in Lausanne. Together we then went out to the people in Basel to find out about their attitude towards this scientific discipline. In our survey, we found that most of the people we asked had either not heard about synthetic biology before or in case they had, they did not feel infomred enough. It seems that the idea of genetically modified organisms is something that causes a lot of discomfort in many people. You can find out more about the results of our survey on our Awareness page. Despite Switzerland being a small country, there is a big diversity amongst different parts of the population. Therefore it was very interesting for us to compare the opinions we got from people in Lausanne, one of the bigger cities in the French-speaking and generally more liberal part of Switzerland, to the answers we received in Basel, located in the German-speaking part of Switzerland.
Have you ever heard of the term 'Synthetic Biology'?

Lausanne

Basel
According to the given definition and examples, what do you think is the public attitude towards this scientific discipline?

Lausanne

Basel
Do you think there is sufficient interaction between Scientists and the general public in Switzerland?

Lausanne

Basel
Colombia
We helped Colombia with the debugging of their transformation protocol and supported them by sharing our experiences in the synbio workflow. We exchanged ideas about how to improve both our systems. Also, we helped them with their modeling in python.