Difference between revisions of "Team:Austin UTexas/Failure Analysis"
| Line 19: | Line 19: | ||
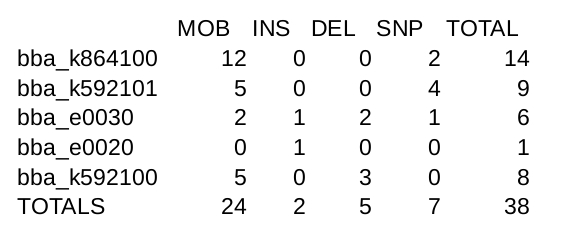
[[Image:2015_UT_AUSTIN_gdparse_output.jpg|900px|thumb|center| Output of gdparse program, using Genomediff files generated by fri-failure-analysis as input.]] | [[Image:2015_UT_AUSTIN_gdparse_output.jpg|900px|thumb|center| Output of gdparse program, using Genomediff files generated by fri-failure-analysis as input.]] | ||
| − | Combining the two programs, we obtain a set of mutations similar to those gathered through hand analysis. Currently, we still check data by hand to remove some false positives, such as those gathered from poor alignments or low-quality samples. Even so, <b>the programs demonstrate | + | Combining the two programs, we obtain a set of mutations similar to those gathered through hand analysis. Currently, we still check data by hand to remove some false positives, such as those gathered from poor alignments or low-quality samples. Even so, <b>the programs demonstrate substantial efficiency improvements over manual analysis.</b> Additionally, the programs minimize human-introduced errors by enforcing stringent naming constraints on template annotations, sequence files, and part labels. |
=== References === | === References === | ||
Revision as of 01:32, 19 September 2015
A Computational Approach: fri-failure-analysis and gdparse
fri-failure-analysis is a computational pipeline that identifies mutations that occur in bacterial plasmids. Each sample read, generated by Sanger sequencing, is aligned to its ancestor strain (template). By identifying homology between samples and mobile elements, the pipeline is able to correctly identify a host of mutations while minimizing false positives. The program streamlines the process of mutation analysis and offers large efficiency gains over human analysis.
The program is divided into phases. The initial phase creates FASTA template files that will be used to create alignments. These each contain two sequences, either a template and a sample, or a sample and a mobile element. The next phase employs the MAFFT program to perform fast pairwise alignments between each sequence. The final phase consists of two steps; first, determine if a sample exhibits homology to a mobile element; then, identify mutations in the sample using this information. These mutations are stored in the Genomediff filetype, which you can read about [http://barricklab.org/twiki/pub/Lab/ToolsBacterialGenomeResequencing/documentation/gd_format.html here.]
gdparse
gdparse is a tool designed to process Genomediff files by counting up mutations and categorizing them. Since the terminal output of fri-failure-analysis is in this format, the program works well as a post-processing step in our "Breaking is Bad" experiment. The program currently outputs a CSV file that tabulates mutations based on their type (e.g. insertion or deletion). Combined with fri-failure-analysis, this program offers further efficiency gains over hand counting of mutations.
This program works by creating dictionaries that map a category (in this case, a BioBrick name) onto other dictionaries that are organized by the type of mutation, the location of the mutation on the reference sequence, or both. Since reference sequences are stored along with each mutation in a Genomediff file, gdparse simply finds the reference sequence in a separate folder and determines what annotation contains the given mutation.
Results
Combining the two programs, we obtain a set of mutations similar to those gathered through hand analysis. Currently, we still check data by hand to remove some false positives, such as those gathered from poor alignments or low-quality samples. Even so, the programs demonstrate substantial efficiency improvements over manual analysis. Additionally, the programs minimize human-introduced errors by enforcing stringent naming constraints on template annotations, sequence files, and part labels.
References
All code maintained by Tyler Camp. fri-failure-analysis and gdparse are released under the GNU General Public License and are free software. Links to the repositories are included below.