Team:Sydney Australia/Parts
Contents
Parts Outline
| Part # | Name | Type | Function | Elements |
|---|---|---|---|---|
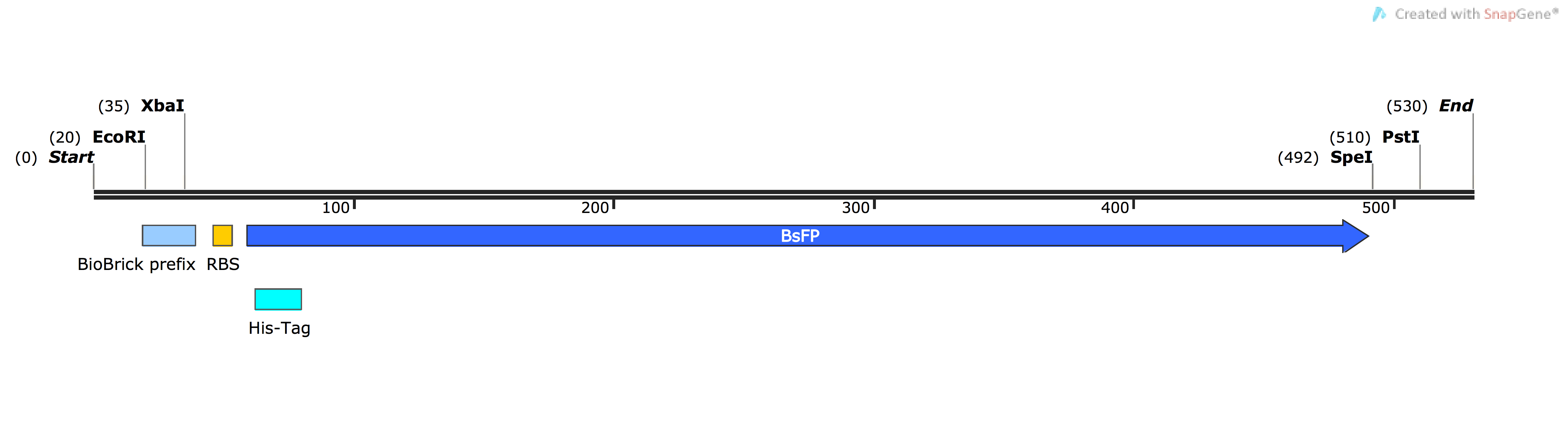
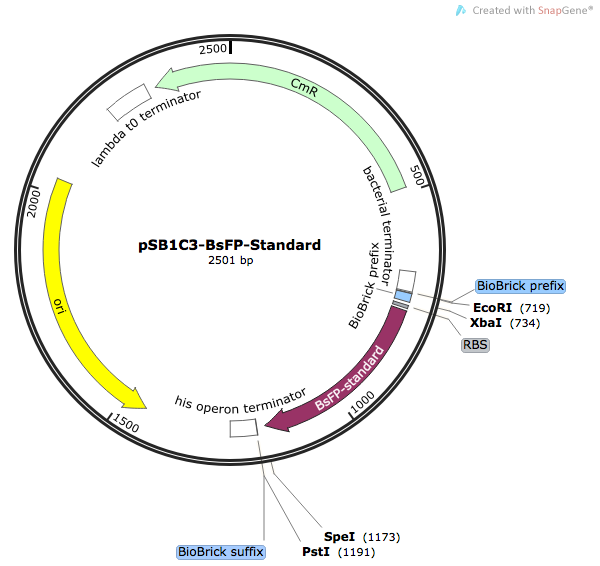
| BBa_K1736000 | pSB1C3-BsFP | Composite | B. subtilis red fluorescent protein primarily used in fluorescence assays as a marker for a range experiments | Harmonised (Spencer et al. 1 method), native B. subtilis RBS & HisTag |
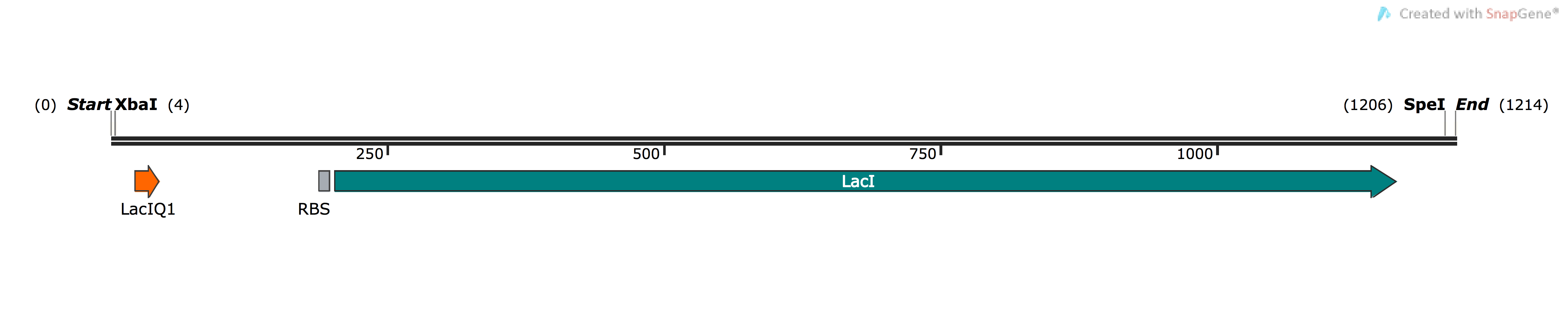
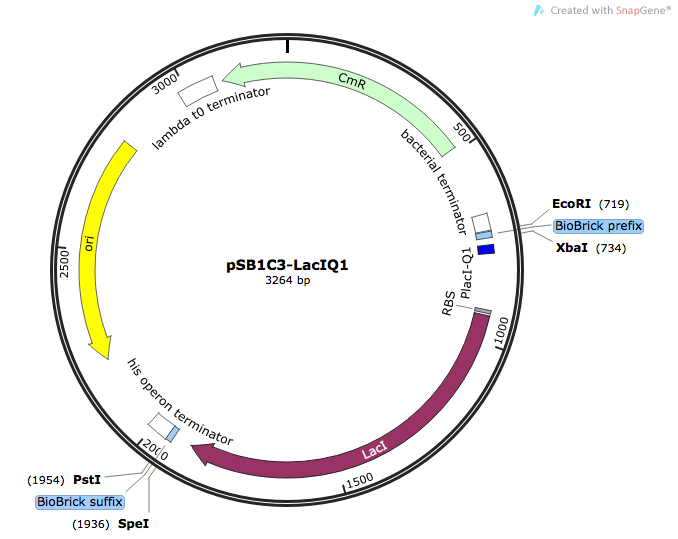
| BBa_K1736300 | pSB1C3-LacI-LacIQ1 | Composite | LacI repressor protein for regulating genes under lac operon expression system | LacIQ1 strong constitutive promoter, E. coli RBS & LacI repressor protein |
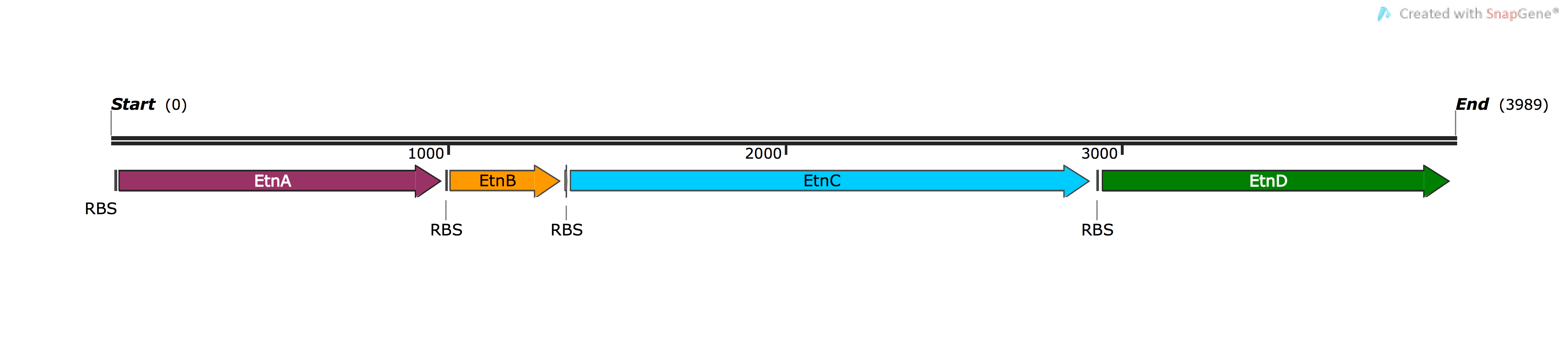
| BBa_K1736200 | pSB1C3-EtnABCD | Composite | Expression of optimised (for P. putida) cytoplasmic di-iron M. chubuense ethene MO, with E. coli RBS | Four ORFs of EtnABCD subunits, native M. chubuense RBS upstream of each ORF |
Characterisation
BBa_K1736000: B. subtilis Fluorescent Protein (BsFP)
The Bacillus subtilis flavin-binding fluorescent protein (BsFbFP, or BsFP) is native to the B. subtilis bacteria, and homologs are also found in several other bacterial genera. This flavin-binding fluoroprotein has a different chromophore to the more widely used GFP family of fluoroproteins, and BsFP has several advantages over the more-traditional fluoroproteins such as GFP ([http://parts.igem.org/Part:BBa_E0040 BBa_E0040]). The BsFP gene is smaller than GFP, and it is capable of folding and fluorescing under anaerobic conditions, unlike GFP and its homologs. The BsFP folds into a fluorescent form faster and more efficiently 1. In this project, we used the BsFP protein to experimentally validate our novel codon harmonisation algorithm, TransOpt. We were inspired to choose this protein as our model system based on the work of previous iGEM teams, who developed related parts [http://parts.igem.org/Part:BBa_K376004 BBa_K376004], [http://parts.igem.org/Part:BBa_K1094000 BBa_K1094000] and [http://parts.igem.org/Part:BBa_K660000 BBa_K660000]. We tried to improve the function of BsFP by utilising three different codon-optimisation / harmonisation approaches; these were our novel 'in-house' algorithm TransOpt, the standard harmonisation methods proposed by Angov et al. 2 and a ‘fast-translating’ method. More information on the difference between these can be found here. Our model hypothesised based on the previous literature that the ribosome translation kinetics profile was influenced by both the copy number of each specific tRNA gene in the host organism, and by the codon redundancy, and that these two factors were paramount in controlling protein folding, which in turn yields protein activity 2. In this experimental validation, we performed fluorescence assays to detect the correctly-folded proteins, with the assumption being that higher fluorescence is characteristic of a better folded protein. We generated the following four sequences:
- BsFP-WT: native sequence from the native host B. subtilis
- BsFP-fast: all codons were replaced with the E.coli versions that possessed the fastest translation rate, as quantised using the approach of Spencer et al 2
- BsFP-standard: harmonised BsFP generated via standard harmonisation using the rate quantisation approach of Spencer et al 2
- BsFP-TransOpt: optimised BsFP sequence generated using our new TransOpt algorithm
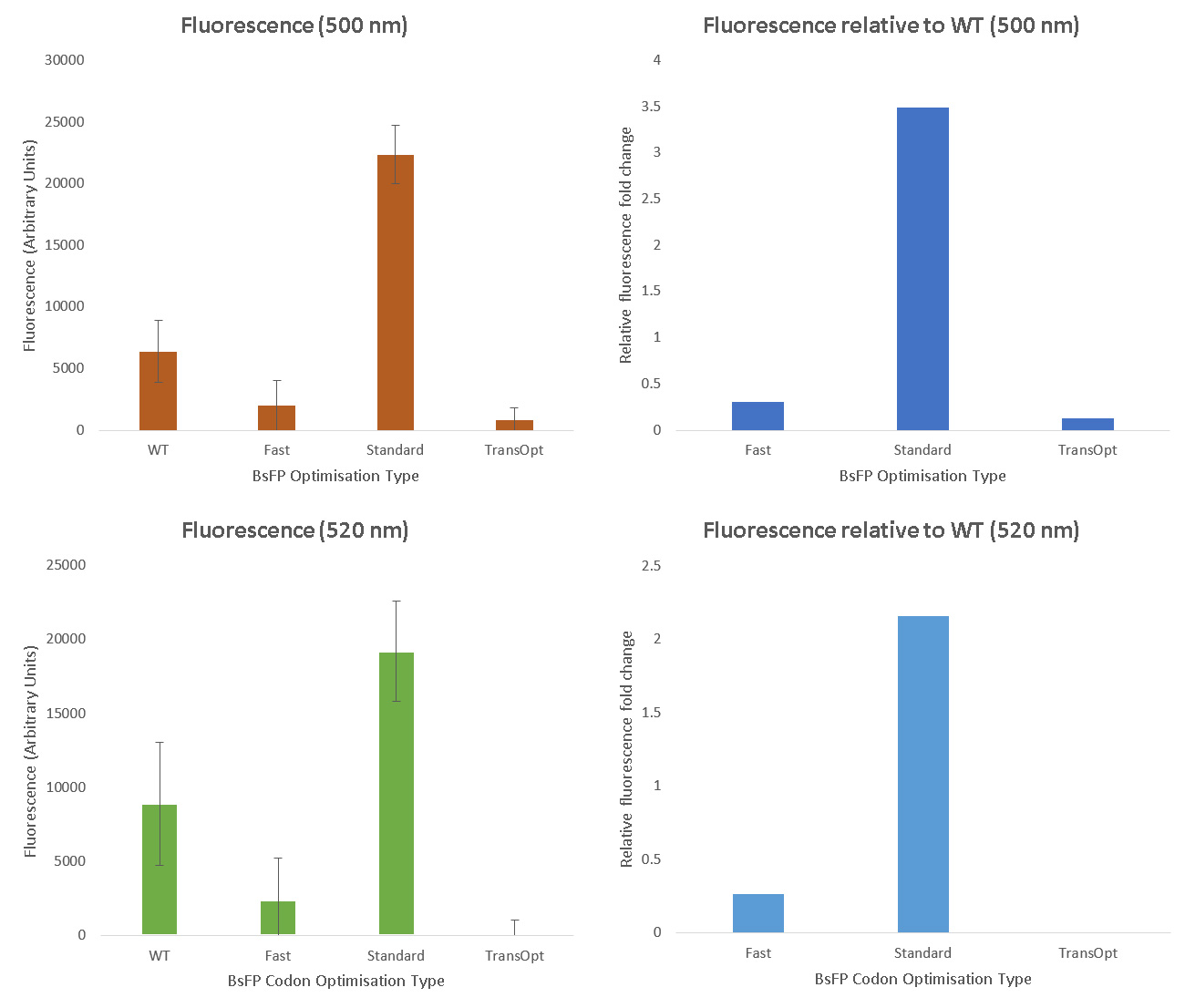
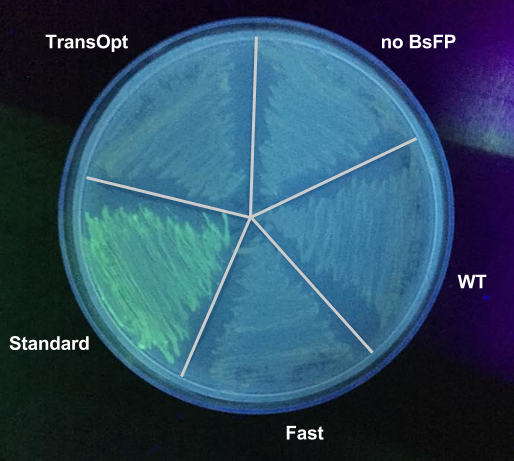
Each variant of BsFP was cloned into a tetracycline-inducible expression vector (pUS212), transformed into E.coli, and clones confirmed to carry the correct constructs were induced with tetracycline, and then both fluorescence (520 nm) and optical density (600 nm) were measured in triplicate samples of each culture. The fluorescence data were normalised by dividing by the optical density. After performing the fluorescence assay, we found that the BsFP subjected to standard harmonisation ([http://parts.igem.org/Part:BBa_K1736000 BBa_K1736000]) possessed the highest fluorescence, which was over 2 fold higher than the wild-type sequence. The fast-folding and Trans-Opt versions of the gene were less fluorescent than wild type. These data were also qualitatively confirmed by exposing tet-induced patches on plates to a long-wave UV lamp; only the standard-harmonisation clones yielded fluorescence visible with the naked eye.
Although we failed to demonstrate that the TransOpt algorithm facilitated greater fluorescence in the case of BsFP, we predict that this approach may still be useful for other proteins. Due to its poor fluorescence, we did not submit the TransOpt-harmonised BsFP as a Part. Instead, we submitted the standard-harmonisation BsFP gene to the Registry. This Part is a versatile and widely-applicable alternative to GFP type fluoroproteins, due to its high fluorescence, small size, fast maturation, ad oxygen-independence.
This new BsFP Part has the potential to enhance reporter gene assays, improve biomarker tracking in cell tissues, create fluorescent fusion proteins, and many other applications. It is also appropriate to note that the gene encoding the protein is substantially different to the previously submitted parts BBa_K376004, BBa_K1094000 and BBa_K660000; while these cannot be directly compared in terms of fluorescence intensity, we note that our part is the most rigorously characterised of these. In particular, the ability to easily detect its fluorescence on agar plates is especially convenient.
BBa_K1736300: LacI-LacIQ1
As explained in the project description & results page, the LacI-LacIQ1 was a late addition to the project to tackle the issue of toxicity to the expressing cells as a results of overexpression or over-synthesis of the enzymatic products. In this way, we add a layer of control to the expression of our ethene MO enzymes to ensure that they are expressing at appropriate and safe levels. Furthermore, its inactivation during the lab preparatory procedures of transformation and developing competent cells can aid the cell to deal with the immense amount of physiological stress imposed upon it and avoid an impending apoptosis.
In order to ensure that we have a constitutively strong expression and activity of LacI, we placed the LacIQ1 promoter upstream of the E. coli LacI ORF, which is highly homologous and similar across bacteria. The following parts are already deposited for LacIQ1: [http://parts.igem.org/Part:BBa_K091112:Design BBa_K091112] and [http://parts.igem.org/Part:BBa_K091131 BBa_K091131]. As for LacI, many parts for the gene have been deposited such as [http://parts.igem.org/Part:BBa_C0012 BBa_C0012]. However, there is no composite part where the LacI gene and LacIQ1 have been placed together, making a new addition to the registry which will be useful for future and teams and experiments relying on strong expression control of gene under lac operon regulation.
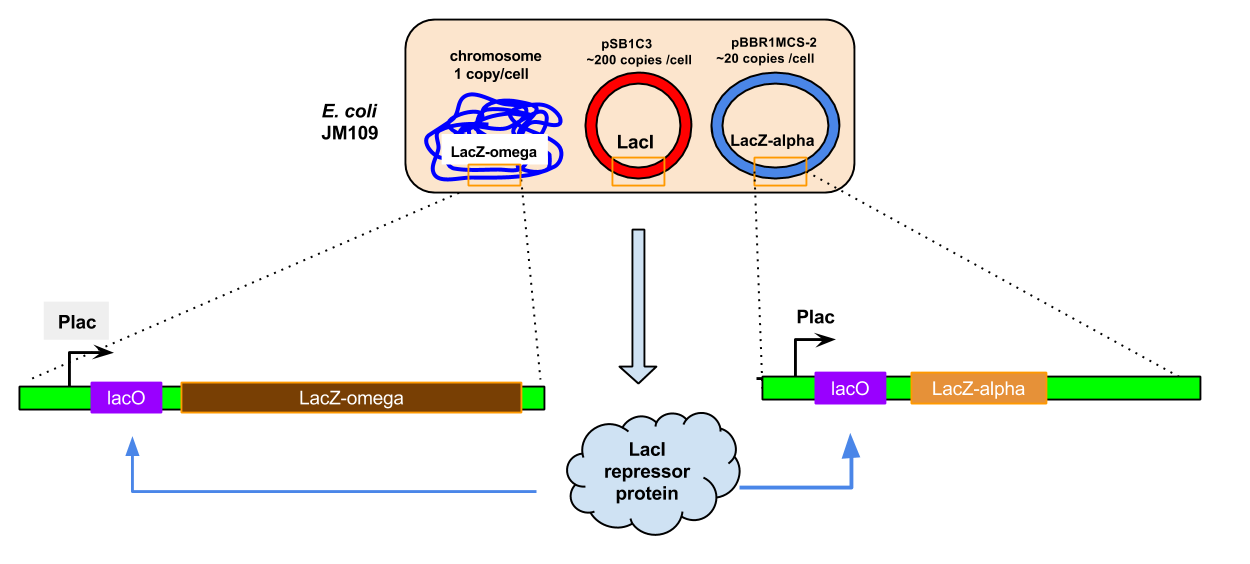
To confirm that the composite part is functional, we ran an experiment to qualitative check for the activity of LacI via the reporter gene LacZ and it's hydrolysis of X-gal (derivative of lactose which turns blue upon hydrolysis by B-galatosidase encoded by LacZ-alpha, and is not taken up by the bacteria). E. coli competent cells containing the LacZ-alpha gene (placed under the control of lac promoter) were transformed with LacI-LacIQ1 and placed in media containing IPTG and no IPTG to induce expression of LacZ (B-galactosidase), with both having equal amounts of X-gal.
After the induction, there were blue colonies on the IPTG media as expected, however, some blue colonies (to a lesser extent qualitatively) in the media containing no IPTG. This is unexpected and hypothesised to be due to the genomic native host lac operon system being responsible for the B-galactisidase activity and expression. This explanation becomes more credible considering that the host contains a lac operon containing both fragments of B-galactosidase enzyme LacZ-alpha and LacZ-beta, where each are under the control of a separate lac promoter. Hence, due to the high-copy presence of LacZ-alpha in a recombiannt vector in the cells and the higher expression of repressor proteins (also due to the high copy number of recombinant vectors containing LacI-LacIQ1), most of the repressor protein block the LacZ-alpha on the vectors, not the genomic ones. Furthermore, it could also be possible that the LacI repressor cannot control the genomic LacZ-beta expression, which can still be expressed and perform X-gal hydrolysis. Hence, while we know this system should work due to countless studies including a paper on LacIQ1 by Glascock CB et al. 3, every user of this construct has to check for the absence of LacZ subunits in the genome which could breakaway from the claws of LacI repressor control.
BBa_K1736200: Ethene MO (EtnABCD)
This construct was the main component of the project which would be responsible for the expression of ethene MO enzyme and the biosynthesis of ethene oxide from ethene. After performing successful Golden Gate cloning (putting 3 Gblocks into a vector, you can find more information about our method here), we transformed it into P. putida competent cells and observed no ethene oxide after incubating the sample in the presence of ethene through utilising the qualitative NBP assay. We believed that the constitutive expression of ethene MO straight after transformation would put too much stress on the cell, hence, we introduced LacI to control the expression of the system. However, after IPTG induction, the NBP assay failed to detect any traces of ethene oxide, which shows that the inactivity lies within the enzyme itself, not the cells. One reason for the ethene MO inactivity could be that our inhouse harmonisation tool TransOpt is not able to yield active and properly folded proteins (through changing the codon sequences) as was found in the experimental validation with BsFP (for more information on the experimental validation, visit results and discussion). This conclusion on the non-functionality of the genetically modified P. putida host due to the ethene MO inactivity is further supported by the fact that the previous ethene MO constructs came positive and the NBP assay was reliable when tested with epoxides.
While this particular construct and sequence of ethene MO was not functional, future iGEM teams should consider the method used and the harmonisation considerations taken into account for any future efforts focused on the development of a functional ethene MO.
Important notes concerning this part:
- pSB1C3-EtnABCD part submitted is missing the XbaI cut site at the BioBirck prefix.
- A mutation (D91E) exists in the sequence due to accidental change while removed BsaI cut sites, however, the change is conservative and is present in ethene MO of other Mycobacterium strains, which means that it is unlikely to have had an effect on the function of the enzyme.
References
1 Mukherjee, A., et al., Characterization of flavin-based fluorescent proteins: an emerging class of fluorescent reporters. PLoS One, 2013. 8(5): p. e64753.
2 Angov E et al., 2008, “Heterologous Protein Expression Is Enhanced by Harmonizing the Codon Usage Frequencies of the Target Gene with those of the Expression Host”, PLoS One, Vol 3.
3 Glascock CB. et al. Using chromosomal lacIQ1 to control expression of genes on high-copy-number plasmids in Escherichia coli. Gene. 1998 Nov 26;223(1-2):221-31.