Team:Austin UTexas/Project/Strain Study
Comparing Genetic Device Stability in Different Strains
Assessing a Yellow Fluorescent Protein Gene for Stability
Four strains of E. coli were selected to be transformed with [BIOBRICK NAME&DESCRIPTION HERE]: TOP10, MDS42, BL21 (DE3), and BW25113. These strains were all selected because of the fact that they are commonly used strains in synthetic biology research. The MDS42 strain was selected because is has had all characterized IS elements removed from its genome (Umenhoffer et al. 2010). Our earlier experiments found that a major cause of mutation in genetic devices, and specifically in this genetic device, is transposable elements inserting into the genetic device's sequence. So, our hypothesis is that cultures of MDS42 cells with [SYFP - BIOBRICK NAME] will exhibit greater fluorescence longevity than the three other strains.
[Do we want to mention the other YFP plasmids? Since you only present data from one of the plasmids...]
All four strains were transformed with three plasmids that were designed and constructed in spring of 2015. The plasmids contained the same backbone (pSB1C3) and the same medium promoter and ribosome binding site. The three plasmids differed only in the type of yellow fluorescent protein used: Yellow Fluorescent Protein, Super Yellow Fluorescent Protein, and Enhanced Yellow Fluorescent Protein [Biobrick names? link?]. These three were selected because of the variance in stability patterns they expressed in our initial experiments. After each strain was transformed with each plasmid, they were grown in LB overnight and then preserved as glycerol stocks.
To determine the relative stability of thee Super-Yellow Fluorescent Protein genetic device in each of the four strains, six replicates of each strain containing the genetic device were grown in LB media. There were 24 cultures in all. After each culture was grown overnight, it was dubbed "Day 1". A Day 1 culture contains approximately 35 generations of E. coli cells.
A sample of Day 1 culture was used to carry forward to the next day via a 1:1000 dilution, yielding an additional 10 generations on Day 2. A sample of Day 1 culture was also taken to measure fluorescence using flow cytometry. The remaining culture was used to create frozen stock for the future. This frozen stock was kept in case we needed to make plasmid preps or go back to the culture on a specific day.
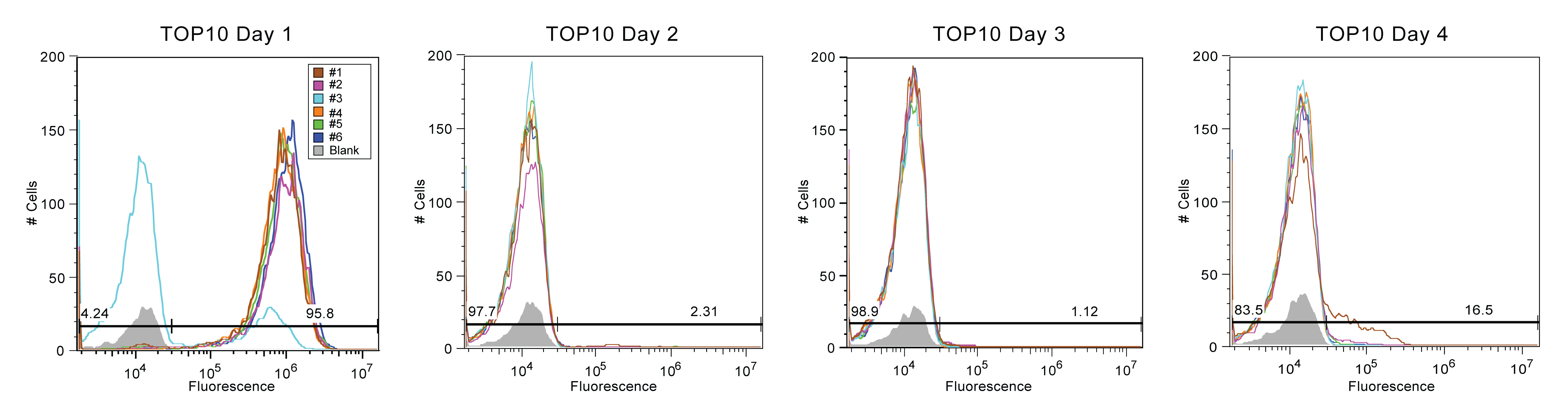
[Description of flow cytometry... move to somewhere else? This seems like procedure] Re-suspension of each culture using PBS preceded the use of the flow cytometer. Each sample was then pipetted into a well in a 96-well plate, with every six samples separated by a well filled with PBS only. The flow cytometer reads to fluorescent value of each well by sipping each well automatically using a syringe. The media flows from the syringe and into cytometer to be passed through a laser which counts the number of cells (called events) and the intensity of the fluorescence and repeats this for each filled well. The first three days of samples that were read using flow cytometry are seen in Figure 1. The x-axis is the magnitude of fluorescence, which is using a logarithmic scale. The y-axis is the count of cells or objects in sample with a particular fluorescence.
[Results! Move to somewhere else?] By looking at the counts of positive fluorescence from day to day, it is clear that the Top10 strain is quickly breaking down, while the MDS strain has remained stable. After four days, the Top10 group of SYFP appeared to have a population that was mostly none fluorescent (See Figure 2). On the sixth day, all of the MDS42 cultures, the third and fifth BL-21 cultures, and the second BW-25113 culture were carried forward and recorded using a flow cytometer.
[Keep this here? Ask Professor Barrick] Our project this year explored the genetic stability of fluorescent protein devices and potential mutational variance between different strains of E. coli. In particular, MDS42, a strain of E. coli without transposons, we hoped to observe mutants that otherwise would have masked by the frequency and prevalence of mutations due to IS (insertion sequence) elements.
Procedure
To continue studying evolutionary stability, we began a variation of our spring experiment. Instead of studying only Top10 E. coli, we expanded our experiment to include other common strains, specifically: BL-21 (DE3), BW-25113, and MDS-42. The MDS-42 cells had the added benefit of containing only a minimal genome.
Before propagating the cell cultures, for each strain, we streaked transformed cells from frozen stocks on to an LB/CAM plate and let them incubate overnight. The next day, we chose six indepedent and fluorescent colonies for each strain. To ensure all cells were retrieved for a more accurate generation time, we pierced the full depth of the agar with a micropipettor tip large enough to encompass the entire colony. We then placed the colony in 10 mL of LB/CAM media and grew all 24 cultures overnight in the shaker at 37° C and 215 RPM. We refer to these cultures as 'Day 1' in our results. The next day, we began the propagation/mutation* phase of our experiment.
From this point forward, each morning we retrieved the overnight cultures from the shaker and checked for fluorescence using a blue light, recording any observations. Then, we would vortex each culture to homogenize the liquid, we used 10 μl to start a fresh 10 mL overnight culture with LB/CAM media. Next, we froze 3 mL of culture in 15% glycerol at -80° C for storage and spun down 4.5 mL of culture for minipreps. In the spring, we used a dark reader to determine a single fluorescence value. However, over the summer we switched to using flow cytometry as a more accurate measure of fluorescence. So, with the remaining 2 mL of culture, every few days we would use the flow cytometer to determine the proportion of cells which were still fluorescent.
After a culture appeared to stabilize at either a complete or partial loss of fluorescence, we stopped pushing the culture forward.
Results
Although we are still awaiting sequencing results, data from the flow cytometer helps to elucidate strain to strain differences in stability.
Flow data for the Top10 cultures corroborate findings from the spring--fluorescence diminishes quickly and precipitously. Max fluorescent cell intensity typically centers around 10^6, while our control media (with zero fluorescence) centers around 10^4. By the second day, the fluorescence cell populations were almost completely absent and on the fourth day, we stopped pushing Top10 forward.
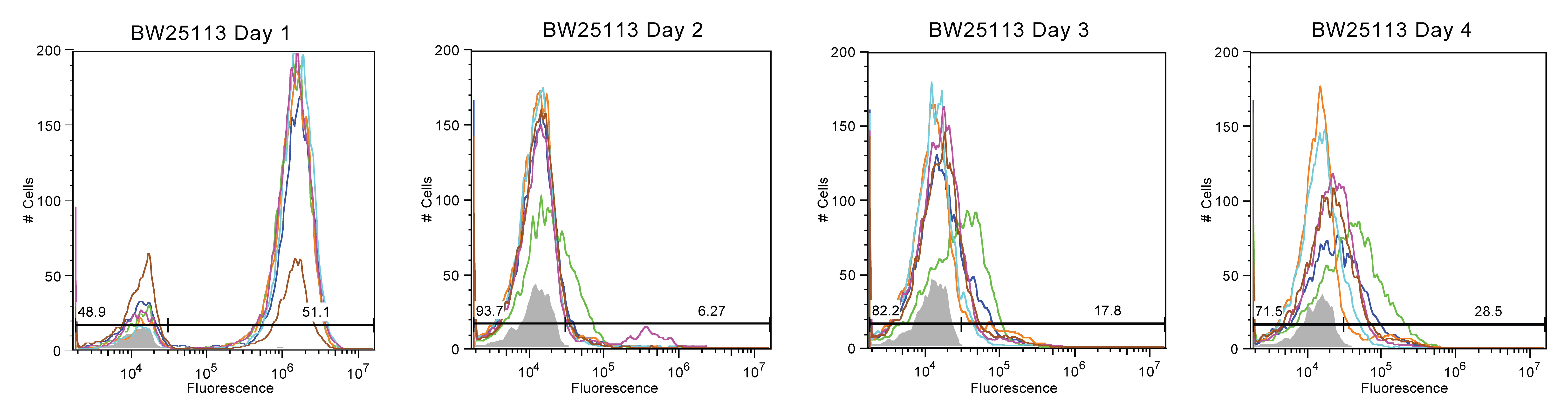
BW-25113 behaved similarly, on the second day we consider lessthan 20% of the cells to be still fluorescing. However, several of the samples appeared to maintain a small proportion of the cells (20%) with reduced (~4*10^4) fluorescence.
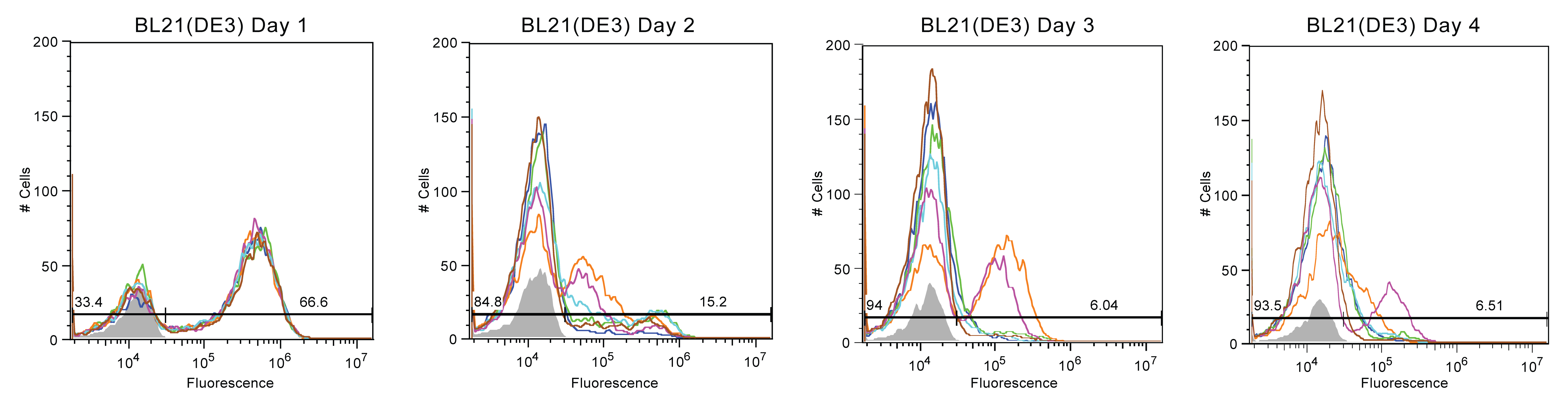
Most BL-21 (DE3) cultures experienced a similar, drastic reduction in fluorescence. However, 2 of the 6 in these strains maintained a small fluorescent population, at approximately 10^5, up to day 3, with one persisting until Day 4.
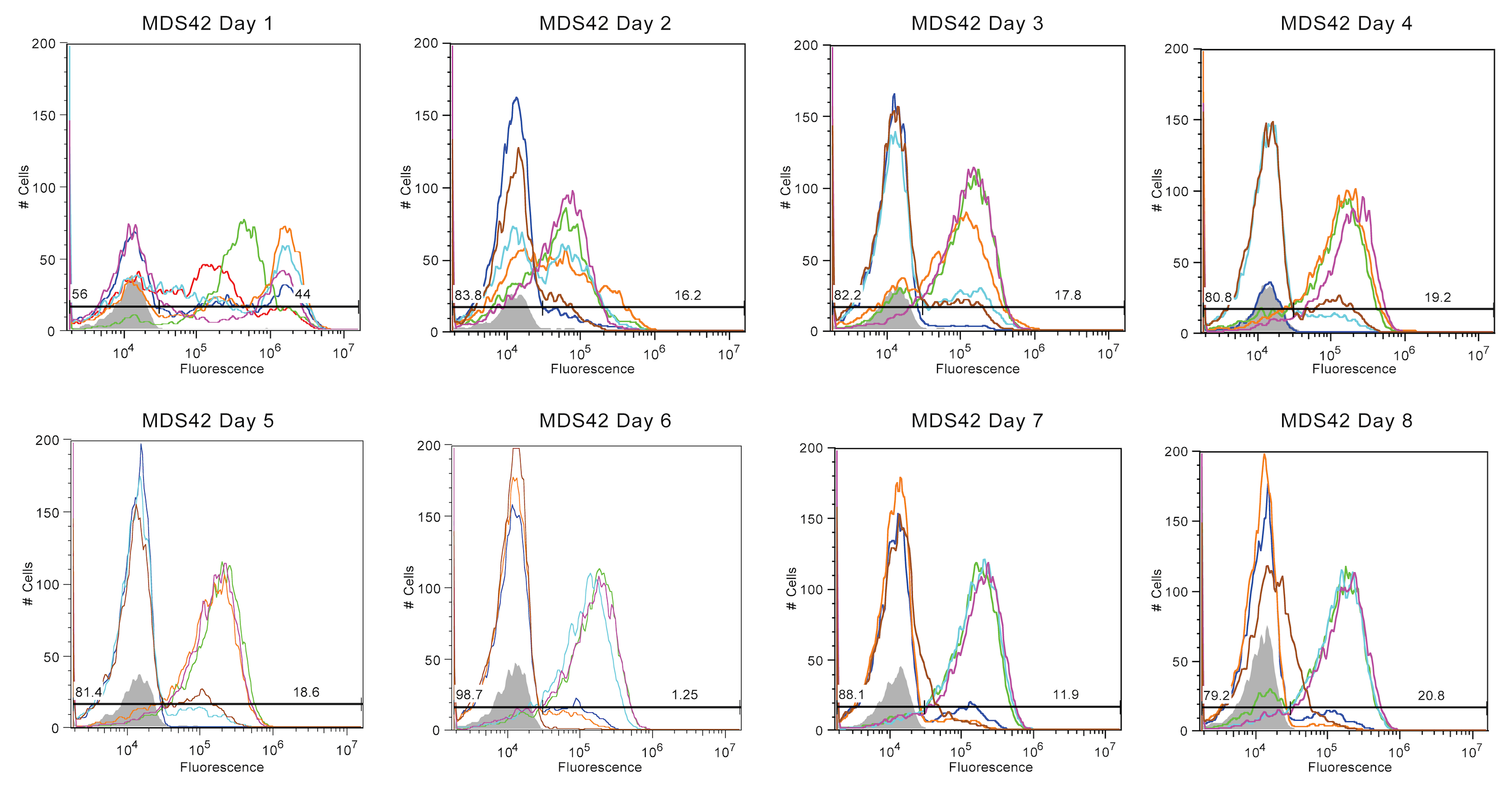
Finally, fluorescence in MDS-42 cultures endured the longest. Three of the samples had lost fluorescence by Day 3, while the remaining three samples maintained a reduced fluorescence level (~2*10^5) until Day 8.
In summary, Top10 cells were most unstable, with nearly all fluorescence being lost by Day 2. In contrast, MDS-42 cultures exhibted the best stability but also the most variance, with some cultures breaking after only three days and some enduring for an additional five days. BW-25113 and BL-21 (DE3) samples experienced moderate fluorescence stability, with trends less extreme than Top10 or MDS-42.
Discussion
Overall, MDS-42 was best able to maintain fluorescence throughout the duration of the experiment. We speculate that, without the very advantageous mutant that arises from transposons, other mutations were able to compete and resulted in moderate maintenance. These mutations may not have directly affected the SYFP2 coding sequence. For instance, auspicious mutations could have occurred in the genome, or other factors such as the plasmid copy number or antibiotic resistance gene could have been altered. Future analysis of the sequencing data will further illuminate what caused the trends seen in these graphs.
References
- Umenhoffer, Kinga et al. “Reduced Evolvability of Escherichia Coli MDS42, an IS-Less Cellular Chassis for Molecular and Synthetic Biology Applications.” Microbial Cell Factories 9 (2010): 38. PMC. Web. 18 Sept. 2015.