Team:Austin UTexas/Project/Caffeine
Expanding on Caffeinated Coli
Background
The ∆guaB strain lacks the guaB gene, which is required for the de novo biosynthesis of guanine. However, when the ∆guaB strain contains the pDCAF3 plasmid, the strain can still make guanine if there is xanthine present OR if there are methylxanthines that can be demethylated to xanthine. Thus, we can estimate the amount of caffeine in a solution by measuring the growth of a ∆guaB strain with pDCAF3 and then comparing the amount of growth to a standard growth curve using the exact same strain but in solutions with known amounts of caffeine.[1]
While this works well for solutions that only contain caffeine, the ΔguaB pDCAF3 bacteria will grow in the presence of any methylxanthine, xanthine, or guanine. Many of these are present in drinks made from organic products, such as coffee or tea, where knowing the exact amount of caffeine would be helpful. Thus, our goal this summer was to create a set of plasmids based on the pDCAF3 plasmid that will allow us to subtract out the background growth from those non-caffeine molecules.
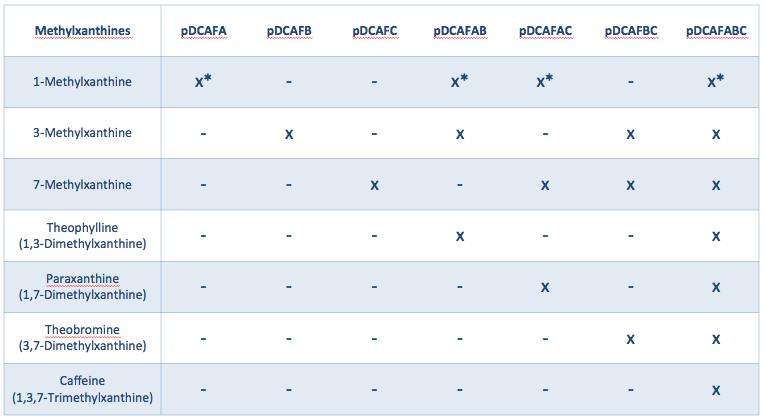
The pDCAF3 plasmid contains five genes that allow it to demethylate caffeine: three (ndmA, ndmB, and ndmC) that specifically demethylate each of the three methyl groups, and two (ndmD and gst9) that are necessary for the other three to work, but don’t contribute directly to the demethylation. We decided to create a set of seven plasmids that all contain ndmD and gst9, and either contain one of the other three, a pair of the other three, or all of the other three genes (Figure 1). Without all three of ndmA, ndmB, and ndmC, the bacteria will only be able to degrade some methylxanthines, not all of them, and only having all three will allow them to degrade caffeine specifically. To get a much more accurate measurement of the amount of caffeine in an organic sample, we can grow parallel cultures containing a single sample of coffee, tea, or some other drink. The only difference between the parallel cultures is which plasmid from figure 1 (in a ∆guaB strain) is present. Once growth is complete, we can subtract out growth derived from non-caffeine molecules (as determined in the parallel cultures that can not degrade caffeine) from the single culture that can degrade caffeine.
Plasmid Design
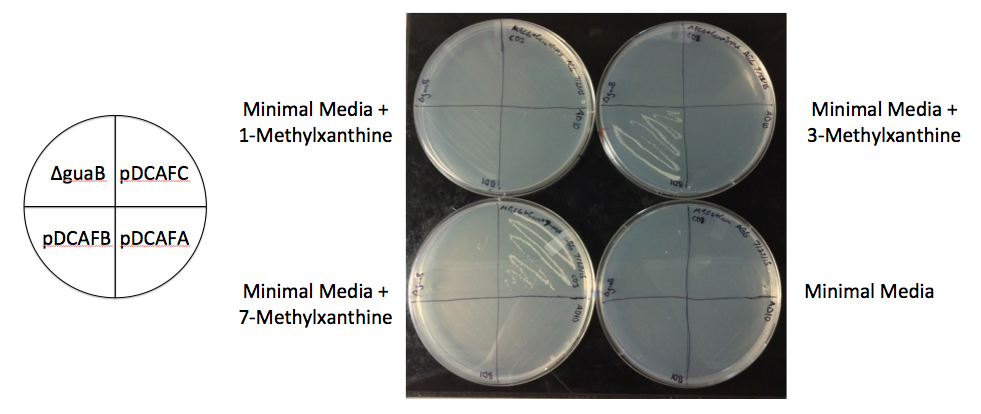
We created a set of seven plasmids containing subsets of the ndmA, ndmB, and ndmC genes that should each degrade and grow on a different subset of methylxanthines (Figures 1, 2). The plasmids were named based on the pDCAF3 naming scheme and which subset of genes it contains (e.g. pDCAFC contains ndmC, ndmD, and gst9, while pDCAFAC contains ndmA, ndmC, ndmD, and gst9). In addition, each gene was giving a different RBS (see Increased Stability below). The genes were amplified from the pDCAF3 plasmid and cloned using the BioBrick Standard Assembly method. Once each plasmid was completed, they were transformed into ∆guaB E. coli and grown on minimal media supplemented with the highest order methylxanthine they can degrade in order to prevent the genes from breaking.
Detection of Caffeine and Related Molecules
This set of plasmids can be used to measure the concentration of methylxanthines in solutions, even caffeinated beverages like coffee or tea that have a mixture of methylxanthines. In order to do so, we first had to create standard growth curves of the strains of ∆guaB E. coli containing the different plasmids. We grew each strain in triplicate in minimal media cultures containing different concentrations of the methylxanthine they degrade. These cultures were grown overnight and measured for OD600 the next day. The results were graphed against the concentration of methylxanthine in the media, and a fit was made for the cultures over the linear range for the xanthine added, with the cutoff being around 100 µM (Figure 4). Once this growth curve is established, the strain can be grown in media containing methylxanthines alone or in a mixture in known or unknown concentrations.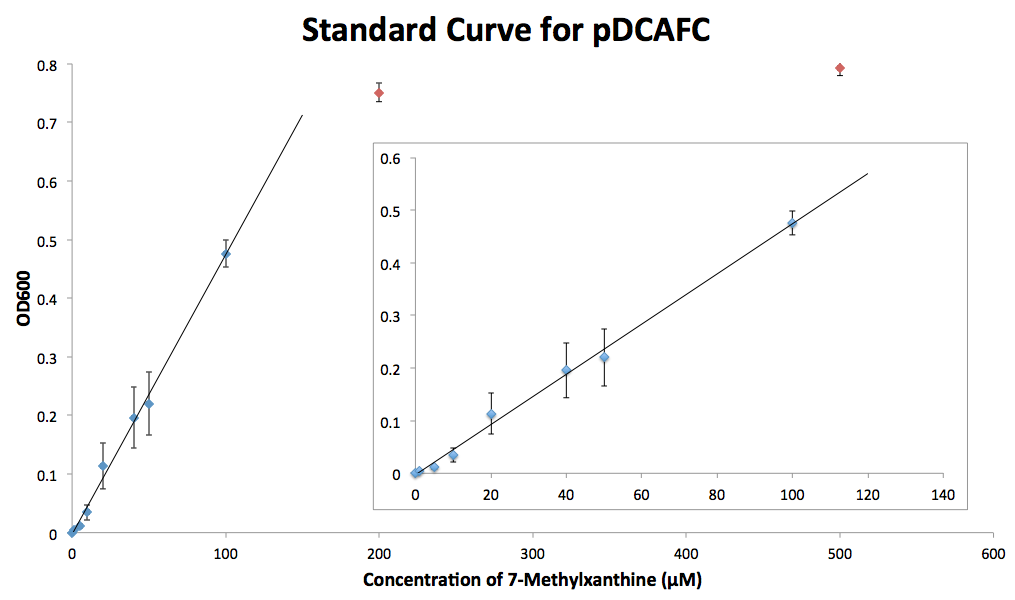
Increased Stability
In addition to creating the six plasmids with combinations of genes that did not exist before, we also redesigned the original pDCAF3 plasmid for greater stability. The pDCAF3 plasmid contained a synthetic operon with the five demethylation genes under the control of one promoter. However, four of the five genes contained the same RBS and surrounding sequence, meaning that there is a 25 bp sequence of DNA that is repeated four times in the operon. This repeated sequence introduces a risk of repeat-mediated deletion, making the plasmid more unstable than it needs to be. As such, during the cloning process we changed the RBS sequences for each of the genes so that they would stay within an order of magnitude in strength, but would have a different enough sequence to alleviate the chances of repeat-mediated deletions. This makes the entire plasmid more stable while retaining the same functionality.
References
1. Quandt, Erik M., et al. "Decaffeination and measurement of caffeine content by addicted Escherichia coli with a refactored N-demethylation operon from Pseudomonas putida CBB5." ACS synthetic biology 2.6 (2013): 301-307.
2. Summers, Ryan M., et al. "Novel, highly specific N-demethylases enable bacteria to live on caffeine and related purine alkaloids." Journal of Bacteriology 194(8) (2012): 2041-2049.