Difference between revisions of "Team:Austin UTexas/Project/Problem"
Kvasudevan (Talk | contribs) |
Kvasudevan (Talk | contribs) |
||
| Line 9: | Line 9: | ||
Previous research has determined several factors that contribute to the breaking of genetic devices, among them being the relative fitness of organisms without the device over those with it. Strongly expressed devices will break faster than less strongly expressed devices, since they have a greater metabolic load on the organism. Similarly, genes that code for large, 'costly' proteins are more likely to break. | Previous research has determined several factors that contribute to the breaking of genetic devices, among them being the relative fitness of organisms without the device over those with it. Strongly expressed devices will break faster than less strongly expressed devices, since they have a greater metabolic load on the organism. Similarly, genes that code for large, 'costly' proteins are more likely to break. | ||
| − | Another major factor that contributes to device stability is the presence of sequence repeats. The <a href="http://barricklab.org/django/efm/">Evolutionary Failure Mode calculator</a> developed by the Barrick lab identifies DNA sequences that are prone to breaking, such as simple sequence repeats and long sequences of a single nucleotide. | + | Another major factor that contributes to device stability is the presence of sequence repeats. The <a href="http://barricklab.org/django/efm/">Evolutionary Failure Mode calculator</a> developed by the Barrick lab identifies DNA sequences that are prone to breaking, such as simple sequence repeats and long sequences of a single nucleotide. The EFM calculator gives sequences a relative instability prediction (RIP) score, and higher RIP scores indicate more sequences where the gene is likely to break. Utilizing this device shows that if a particular sequence contains many short sequence repeats or even large, similar regions, then that sequence risks mutating in later generations. |
=== Project and Motivation === | === Project and Motivation === | ||
| − | + | Our research focuses on monitoring device stability and determining what kinds of sequences and sequence features contribute to device instability. <b>We wish to identify and better characterize sequences that contribute to the relative instability of genetic devices so they can be used to predict device longevity, as well as be taken into account when creating and modifying such devices</b>. In the spring of 2015, UT iGEM team members utilized the EFM calculator to identify sequences within fluorescent protein coding genes that were more likely to break, or mutate. and we used this information to form hypotheses and determine which fluorescent protein coding genes in our inventories are the most and least evolutionary stable. | |
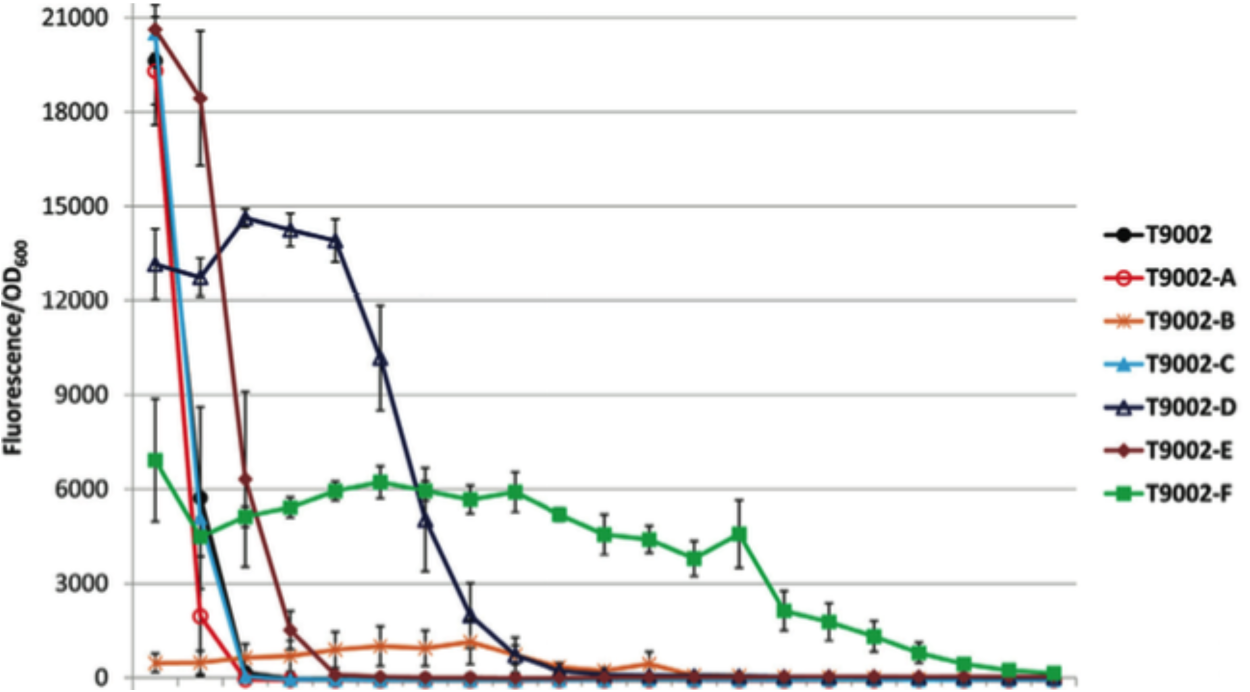
| − | Over the summer, | + | Over the summer, three plasmids were constructed: YFP + Medium RBS; Medium Promoter (BBa_K608006), Super-folder YFP + BBa_K608006, and Enhanced YFP + BBa_K608006. These three plasmids were used to transform four strains of bacteria (Top-10, MDS-42, BL-21 (DE3), BW-25113). We hoped that growing these transformed strains in culture and monitoring their fluorescence and stability would better elucidate the patterns in the sequences that contribute to the instability of genetic devices. Understanding this could lead to the active modification of devices to remove these 'risky' elements, and thus increase the longevity of genetically modified organisms. |
{{Austin_UTexas_Footer}} | {{Austin_UTexas_Footer}} | ||
Revision as of 20:19, 18 September 2015
Problem: Genetic Instability
Background
One of the major issue facing genetic engineering is the overall longevity of genetic devices once inserted into organisms. An organism can be modified, but that does not ensure that the modification will last. Subsequent generations of modified organisms often lose or “break” the genetic device through mutations. Breaking a plasmid or an inserted device decrease the metabolic load of that organism giving it a competitive advantage by allocating cell resources for other tasks. This allows them to dominate a population over some time.
Previous research has determined several factors that contribute to the breaking of genetic devices, among them being the relative fitness of organisms without the device over those with it. Strongly expressed devices will break faster than less strongly expressed devices, since they have a greater metabolic load on the organism. Similarly, genes that code for large, 'costly' proteins are more likely to break.
Another major factor that contributes to device stability is the presence of sequence repeats. The <a href="http://barricklab.org/django/efm/">Evolutionary Failure Mode calculator</a> developed by the Barrick lab identifies DNA sequences that are prone to breaking, such as simple sequence repeats and long sequences of a single nucleotide. The EFM calculator gives sequences a relative instability prediction (RIP) score, and higher RIP scores indicate more sequences where the gene is likely to break. Utilizing this device shows that if a particular sequence contains many short sequence repeats or even large, similar regions, then that sequence risks mutating in later generations.
Project and Motivation
Our research focuses on monitoring device stability and determining what kinds of sequences and sequence features contribute to device instability. We wish to identify and better characterize sequences that contribute to the relative instability of genetic devices so they can be used to predict device longevity, as well as be taken into account when creating and modifying such devices. In the spring of 2015, UT iGEM team members utilized the EFM calculator to identify sequences within fluorescent protein coding genes that were more likely to break, or mutate. and we used this information to form hypotheses and determine which fluorescent protein coding genes in our inventories are the most and least evolutionary stable.
Over the summer, three plasmids were constructed: YFP + Medium RBS; Medium Promoter (BBa_K608006), Super-folder YFP + BBa_K608006, and Enhanced YFP + BBa_K608006. These three plasmids were used to transform four strains of bacteria (Top-10, MDS-42, BL-21 (DE3), BW-25113). We hoped that growing these transformed strains in culture and monitoring their fluorescence and stability would better elucidate the patterns in the sequences that contribute to the instability of genetic devices. Understanding this could lead to the active modification of devices to remove these 'risky' elements, and thus increase the longevity of genetically modified organisms.