Difference between revisions of "Team:Waterloo/Modeling"
(expanding on application section) |
|||
| Line 42: | Line 42: | ||
</div> | </div> | ||
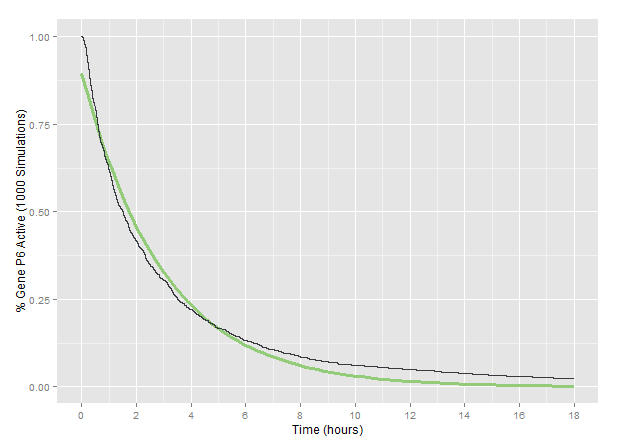
| − | <p>On the <strong>scale of individual genomes</strong>, we used our | + | <p>On the <strong>scale of individual genomes</strong>, we used our <a href="https://2015.igem.org/Team:Waterloo/Modeling/Cas9_Dynamics">dynamics model of CRISPR/Cas9</a> to model viral genomes becoming non-functional over time as frameshift mutations were introduced. The results of running 1000 simulations using our <a href="">sgRNA target design</a> to deactivate the CaMV P6 gene predict the following pattern of P6 functionality over time. |
| − | < | + | <figure> |
| + | <img src="/wiki/images/0/0f/Waterloo_P6_exponential_fit.png" alt="P6 concentration over time with exponential fit" class="img-responsive""/> | ||
| + | <figcaption class="model-caption">Percent of functional P6 genomes observed over 1000 simulations with three targets are shown in black, while an exponential decay fit done with the R nls package is shown in green.</figcaption> | ||
| + | </figure> | ||
| − | <p> | + | <p>The exponential decay fit shown in the graph above was then passed to a model on the scale of <strong>plant cells</strong>. We modelled <a href="https://2015.igem.org/Team:Waterloo/Modeling/CaMV_Replication">CaMV Replication</a> in a single infected cell using ordinary differential equations. The ODE replication model allowed us to see the effect that the CRISPR/Cas9 decay of P6 would have on <strong>virion production</strong>. We also examined the interaction bewteen intracellular plant defenses (RNA interference) and the CRISPR/Cas9 defense and conducted a sensitivity analysis on our parameters.</p> |
<p><strong>Insert graph of virons/time under different conditions</strong></p> | <p><strong>Insert graph of virons/time under different conditions</strong></p> | ||
| − | <p> | + | <p>The ODE model was then scaled up to the level of <strong>plant leaves</strong>. We modeled the spread of infection between plant cells using an agent-based framework, which is described in detail on the <a href="https://2015.igem.org/Team:Waterloo/Modeling/Intercellular_Spread">Intercellular Viral Spread</a> page. The agent-based model allowed us to explore the effect of intercellular defense signalling (which leads plant cells to resist to infection and become suseptible to apoptosis).</p> |
<p><strong>Insert gif of agent-based model with and without P6</strong></p> | <p><strong>Insert gif of agent-based model with and without P6</strong></p> | ||
</section> | </section> | ||
| − | + | <p>Overall, our multi-scale model of CRISPR Plant Defense allowed us to investigate the extent to which we could slow CaMV spread using CRISPR/Cas9 and its interaction with host defense systems such as RNA interference and intercellular defense signalling.</p> | |
| − | + | ||
| − | + | ||
</div> | </div> | ||
</html> | </html> | ||
{{Waterloo_Footer}} | {{Waterloo_Footer}} | ||
Revision as of 01:39, 19 September 2015
Modeling
Mathematical modeling is a core part of Waterloo iGEM: we have nearly as many team members typing furiously away in our dry lab as we do wrangling transformations in our wet lab. This year, we created models in Python, MATLAB and NetLogo that examine the feasibility of our design and provide tools for assessing future designs. The code for each model is available on our GitHub and details on the formulation of each model may be found in the pages linked below.
Cas9 Frameshift Dynamics
Some visual representation of the model and ~100 words about what it contributed to our project, with a link to the CRISPR/Cas9 Frameshift Dynamics page.
PAM Structural Bioinformatics
Some visual representation of the model and ~100 words about what it contributed to our project, with a link to the PAM Structural Bioinformatics page.
Overall, our multi-scale model of CRISPR Plant Defense allowed us to investigate the extent to which we could slow CaMV spread using CRISPR/Cas9 and its interaction with host defense systems such as RNA interference and intercellular defense signalling.