Team:UCLA/Notebook/Spider Silk Genetics/16 July 2015
Contents
7/16/2015
Transformation Results
- MaSp2 3-, 6-mer in BBa_k525998 in BL21 cells have many colonies on the plate.
- May have plated too much. Will reduce plating density in the future when working with the BL21(DE3) chemical competent cells.
- 9-mer in pSB1C3 has very few small colonies.
MaSp2 ICA 12-mer
- Plan to do 12 mer today: 100 uL wases, 5 min ligation time.
- 1 ul of 50 nM initiator used. Initial binding in 45 minutes.
- 5 minute ligation times for each step.
- 50 ng of each DNA species
- 1 uL of 5 uM solution of each relevant cap.
- eluted in 15 uL of 0.01% Tween-20 solution
PCR amplification for MASp2 ICA 12-mer
- Used the following protocol to set up 2x 25 uL reactions.
- Used crude eluate from ICA as template.
- Amplified with post-elution primers
| Volume (uL) | |
|---|---|
| 5x Q5 Buffer | 5 |
| F-03 | 1.25 |
| G-03 | 1.25 |
| dNTPs | 0.5 |
| Template | 0.5 |
| ddH2O | 16.25 |
| Q5 Polymerase | 0.25 |
| Total | 25 |
| 98 C | 30 sec |
| 98 C | 10 sec |
| 66 C | 20 sec |
| 72 C | 35 sec |
| repeat from step 2 | 20 x |
| 72 C | 2 min |
| 12 C | hold |
Results
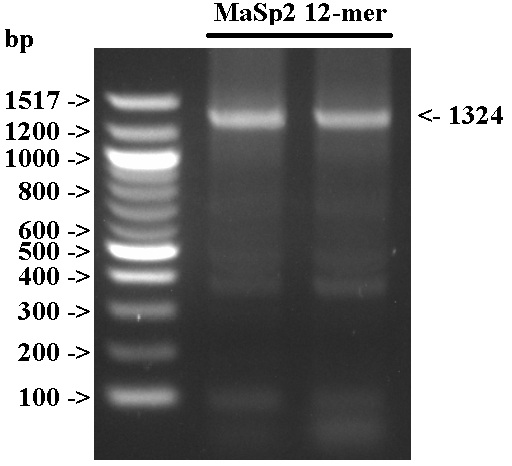
- Cast 1.5% TAE gel to visualize results; used 2 uL of NEB 100 bp ladder.
- We were able to successfully perform ICA to obtain a 12-mer. The accessory bands present may be due to PCR non-specific amplification, or to inaccuracies in the ICA process.
- We excised the 1324 bp band for gel-extraction in preparation for cloning.
Liquid Culture for MaSp2 3-mer, 6-mer, 9-mer
- Grew 3x 5 mL culture for MaSp2 3-mer(T7), MaSp2 6-mer(T7), and 9-mer(pSB1C3).
- Plan to miniprep tomorrow then sequence over the weekend.
- Will have bacterial cultures ready on Monday for the protein expression team to use.