Team:Vilnius-Lithuania/Labjournal
About
pLac+cI.
P10 and P11 plasmids are used. pLac (P11) and repressor cI (P10) ligation.
| Date | Materials | Experiment |
|---|---|---|
| 06 30 | P10 P11 |
P10 and P11 transformation into DH5α. |
| 07 01 | Bacteria inoculation (4 minipreps). | |
| 07 09 | P10 P11 |
Plasmids’ restriction (3 ug each). P10 with EcoRI and XBal (+FastAp). P11 with EcoRI and SpeI. Extraction from gel L10 and L11 are produced. Conc.: L11 – 2,4 ng/μL. |
| 07 13 | P10 |
Repeated restriction of P10 with EcoRI and XBal (+FastAp). Conc.: L10 – 45,3 ng/μL. L10 and L11 ligation. P14 is produced. |
| 07 14 | Bacteria with P14 | Colony PCR of P14 bacteria. 1st and 2nd colonies are inoculated. |
| P14.2 | Sequencing |
Conclusions and Results:
07 14 – all of the colonies are successful, except third colony.
About
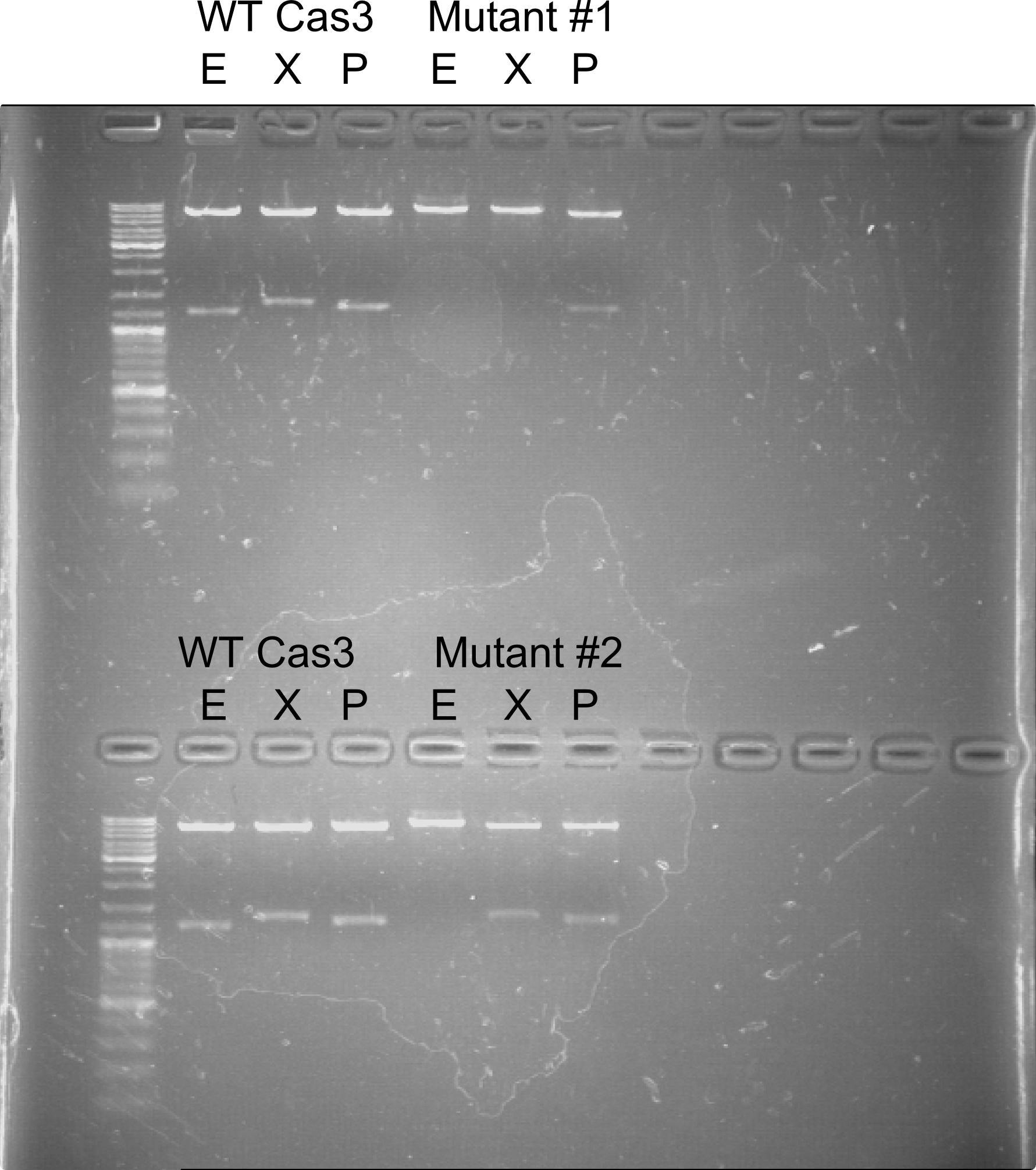
Cas3 with mutated restriction sites.
L7+L15. Concentration - 261 ng/μL
| Date | Materials | Experiment |
|---|---|---|
| 06 30 | P13 P7 |
P13 and P7 transformation into DH5α. |
| 07 01 | Transformants are inoculated (4 minipreps each). | |
| 07 02 | Plasmid extraction. Concentrations: P7 – 174,1 ng/μL and 166 ng/μL. P13 - 216 ng/μL and 131, 5 ng/μL. |
|
| 07 13 | P13 | Cas3 mutagenesis (with ST1/2, ST3/4, ST5/6 primers). |
| 07 16 | Mutated P13 | Cas3 plasmids extraction. |
| 07 21-23 | Mutated P13 | Restriction with EcoRI, XbaI and PstI. |
| 07 27 | Mutated P13 | Cas3 mutagenesis. |
| 07 29 | P7 | Restriction with EcoRI and SpeI. L7 is produced. |
| 07 31 | P13 | |
| 08 02 | Mutated P13 | Mutated Cas3 restriction with EcoRI, XbaI and PstI. |
| 08 05 | Mutated P13 | Cas3 mutagenesis. |
| 08 07 | Mutated P13 | Mutated Cas3 restriction with PstI. Mutagenesis was successful. L15 is produced. |
| 08 11 | L7 L15 |
L7 and L15 ligation with pSB1C3 (EcoRI+XBal+FastAp). P15.1 is produced. |
| P15.1 | Sequencing. |
Conclusions and Results:
07 21 – Mutagenesis was unsuccessful.
08 02 – successfully mutagenized EcoRI and XbaI sites.
08 07 – successful mutagenesis confirmed (XbaI and EcoRI sites are mutated).
08 07 – Successful mutagenesis (PstI, XbaI and EcoRI sites are mutated).
About
P26 – L7+L26 pLux/cI+SRBS
P27 – L7+L27 pLux/cI+MRBS
P28 – L7+L28 pLux/cI+WRBS
P29 – L7+L29 pLux/cI
| Date | Materials | Experiment |
|---|---|---|
| 07 27 |
ST9 ST10 ST11 ST12 ST15 ST16 ST17 ST18 ST19 ST20 |
pLux/cI contruction from oligos – PCR reaction with ST9 and ST10/11/12 primers, then with ST15 and ST16/17/18. Parallel PCR with ST9 and ST19 and then with ST15 and ST20. PCR products purification. |
| 07 28 | PCR products from oligos restriction with EcoRI and SpeI. L26, L27, L28 and L29 are produced. |
|
| 07 28 |
L7 L26 L27 L28 L29 |
Ligation of: L7 and L26, L7 and L27, L7 and L28, L7 and L29 and their transformation into DH5α. |
| 07 29 - - 08 05 |
L7+L26 colony PCR with ST15 and VR. L7+L27 colony PCR with ST15 and VR. L7+L28 colony PCR with ST15 and VR. 1st and 2nd colonies are inoculated. P26, P27, 28 and P29 are produced P26, P27, 28 and P29 restriction with NspI. |
|
| 08 10 | P26, P27, 28 and P29 sequencing with VF2. |
Conclusions and Results:
07 31 – all of the colonies are successful.
08 10 – all biobricks are sequenced succesfully.
About
cI repressor promoter (BBa_R1051) (P4) + screen plasmid intermediate (GFP gene) (BBa_I13504) (P5).
| Date | Materials | Experiment |
|---|---|---|
| 06 30 | P14 | P4 transformation into DH5α. |
| 07 01 | Bacteria inoculation (4 minipreps). | |
| 07 02 | P4 |
Plasmid extraction. Concentrations: P4 - 238,3 ng/μL and 224,3 ng/μL. P4 restriction with EcoRI and XBal. Extraction from gel. L4 is produced (con. 16,2 ng/μL). |
| 07 07 | P5 |
P5 transformation into DH5α. Bacteria inoculation. |
| 07 08 | P5 plasmid extraction (P5.1, P5.2, P5.3, P5.4, P5.5 are produced). | |
| 07 29 | P5.1 and P5.3 |
Restriction with EcoRI and SpeI. Extraction from gel. L5.1A, L5.1B, L5.1C and L5.3A, L5.3B, L5.3C are produced. Conc.: L5.1A – 0,6 ng/μL, L5.1B – 0,6 ng/μL, L5.1C – 2,4 ng/μL, L5.3A – 0,7 ng/μL, L5.3B - 3 ng/μL, L5.3C – 5,1 ng/μL. Ligation of L4 and L5.1, L4 and L5.3. P30 is produced. |
| 08 01 | P30 | Sequencing. |
Conclusions and Results:
07 09 – fragments were not the right ones.
07 29 – the right fragments are produced.
About
P35 - pLux/cI+SRBS(Strong RBS)+Cas3 (L26+L15.1)
P36 - pLux/cI+MRBS(Medium RBS)+Cas3 (L27+L15.1)
P37 - pLux/cI+WRBS(Weak RBS)+Cas3 (L28+L15.1)
P26 – L7+L26 (pLux/cI+SRBS)
P27 - pLux/cI+MRBS(Medium RBS)
P28 - pLux/cI+WRBS(Weak RBS)
| Date | Materials | Experiment |
|---|---|---|
| 07 28 | P15 | Mutated Cas3 with Preffix and Suffix restriction with EcoRI and XbaI. L15 is produced. |
| 07 28 |
P26 P27 P28 |
PCR products from oligos restriction with EcoRI and SpeI. L26, L27, L28 and L29 are produced. |
| 07 28 |
L15 L26 L27 L28 |
Ligation of: L15 and L26, L15 and L27, L15 and L28, and their transformation into DH5α. |
| 07 29 - - 08 05 |
L7+L26 colony PCR with ST15 and VR. L7+L27 colony PCR with ST15 and VR L7+L28 colony PCR with ST15 and VR 1st and 2nd colonies are inoculated. P35, P36 and P37 are produced P35, P36, P37 restriction with NspI. |
|
| 08 10 | P35, P36, P37 sequencing. |
Conclusions and Results:
07 31 – all of the colonies are successful.
08 10 – all biobricks are sequenced succesfully.
About
Mutated Cascade protein.
| Date | Materials | Experiment |
|---|---|---|
| 06 30 | Cd | Cd transformation into DH5α. |
| 07 01 | Transformants are inoculated (4 minipreps each). | |
| 07 02 | Plasmid extraction. Conc.: 329 ng/μL and 294 ng/μL. | |
| 07 14 | Cd | Cd mutagenesis (with ST13/14 primers). |
| 07 16 - 17 | Mutated plasmids extraction. Restriction with EcoRI. | |
| 07 19 | Mutated Cd | Restriction repeat. |
| 07 21 | Mutated Cd | Repeated mutagenesis (with ST13/14). |
| 07 27 | Mutated Cd | Mutated plasmids extraction. Mutated Cd restriction. |
| 08 10 | Mutated Cd L7 |
PCR with mutated Cd with ST26/27. PCR product purification. Conc.: 63 ng/μL. Restriction with EcoRI and SpeI. Mutated Cd ligation with L7. Cd1 is produced. |
| Cd1 | Sequencing. |
Conclusions and Results:
07 16-17 – Mutagenesis was unsuccessful.
07 27 – Mutagenesis conformed as a successful.
About
26-Cd - Cascade with SRBS
27-Cd - Cascade with MRBS
28-Cd - Cascade with WRBS
| Date | Materials | Experiment |
|---|---|---|
| 08 15 | Cd1 | Mutated Cd with Preffix and Suffix restriction with EcoRI and XbaI. L35 is produced. |
| 08 15 |
P26 P27 P28 |
PCR products from oligos restriction with EcoRI and SpeI. L26, L27, L28 and L29 are produced. |
| 08 15 |
L35 L26 L27 L28 |
Ligation of: L35 and L26, L35 and L27, L35 and L28, and their transformation into DH5α. |
| 08 17 - - 08 20 |
L35+L26 colony PCR with ST15 and VR. L35+L27 colony PCR with ST15 and VR L35+L28 colony PCR with ST15 and VR 1st and 2nd colonies are inoculated. 26-Cd, 27-Cd and 28-Cd are produced. 26-Cd, 27-Cd and 28-Cd restriction with NspI. |
|
| 08 21 | 26-Cd, 27-Cd and 28-Cd sequencing. |
Conclusions and Results:
07 31 – all of the colonies are successful.
08 10 – all biobricks are sequenced succesfully.
About
pLac-cI repressor+ pLux/Ci-LuxI
| Date | Materials | Experiment |
|---|---|---|
| 07 19 | pLac (R0010) cI lambda repressor (P0151) |
pLac biobrick was cleaved with EcoRI and SpeI, L10 was constructed. cI Lambda repressor biobrick was digested with EcoRI and XbaI + FastAP, L11 was constructed. |
| 07 19 | L10 L11 |
L11 and L10 were ligated and transformed to DH5α. |
| 07 20 |
Colony PCR was performed with VF2 and VR primers. 1st and 2nd colonies are inoculated. P18 is produced. |
|
| 07 21 | P14.2 P18 |
P14.2 is digested with EcoRI and XbaI + Fast AP. P18 is digested with EcoRI + SpeI. |
| 07 21 | Restriction products are ligated and transformed into DH5α competent cells. | |
| 07 22 |
Colony PCR was performed with VF2 and VR primers. 1st and 2nd colonies are inoculated. P48 is produced and sequenced. |
Conclusions and Results:
07 25 - P48 is sequenced successfully.
About
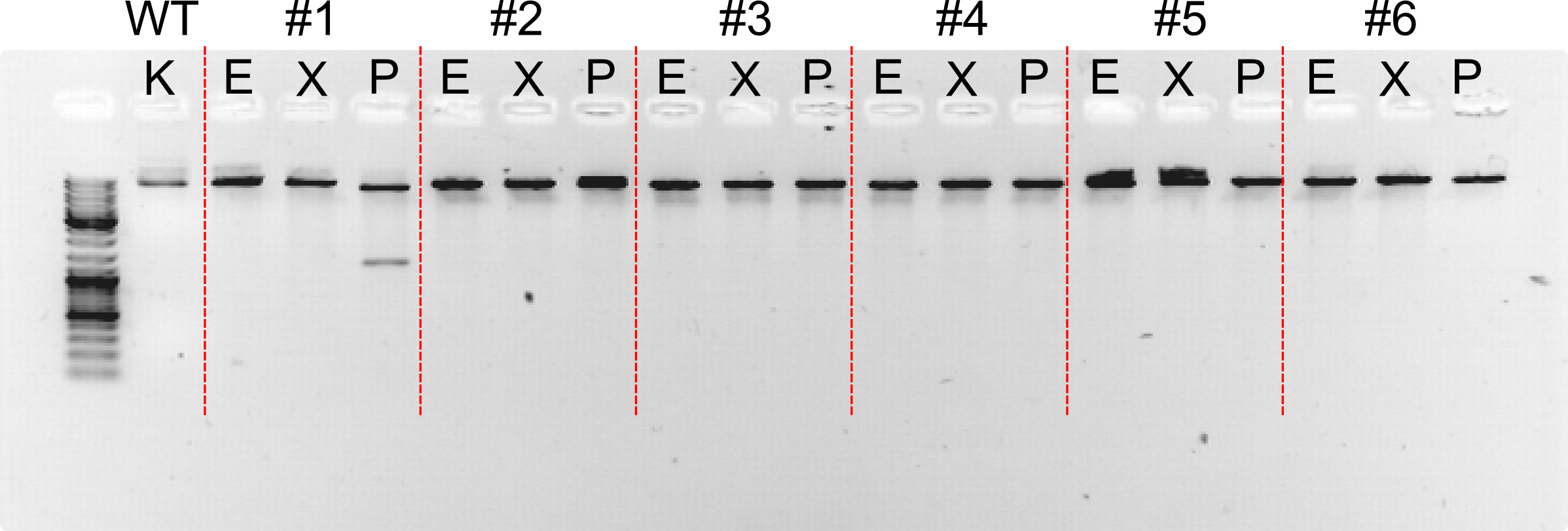
Cd21L26 - Cascade with pLux/cI right promoter and strong RBS
Cd21L27 - Cascade with pLux/cI right promoter and Medium RBS
Cd21L28 - Cascade with pLux/cI right promoter and weak RBS
| Date | Materials | Experiment |
|---|---|---|
| 08 26 |
BL21-DE3 Cd21L26 Cd21L27 Cd21L28 PET-HCsy3 |
Transformation of Cd21L26 and PET-HCsy3 plasmids into BL21-DE3 bacteria. Transformation of Cd21L27 and PET-HCsy3 plasmids into BL21-DE3 bacteria. Transformation of Cd21L28 and PET-HCsy3 plasmids into BL21-DE3 bacteria. |
| 08 27 |
BL21-DE3 Cd21L26 Cd21L27 Cd21L28 PET-HCsy3 |
Transformation of Cd21L26 plasmid into BL21-DE3 bacteria. Transformation of Cd21L27 plasmid into BL21-DE3 bacteria. Transformation of Cd21L28 plasmid into BL21-DE3 bacteria. Transformation of and PET-HCsy3 plasmid into BL21-DE3 bacteria. |
| 08 28 |
BL21-DE3 transformants Cd21L26 Cd21L27 Cd21L28 PET-HCsy3 |
Transformation of Cd21L26 plasmid into BL21-DE3 bacteria (transformed with PET-HCsy3). Transformation of Cd21L27 plasmid into BL21-DE3 bacteria (transformed with PET-HCsy3). Transformation of Cd21L28 plasmid into BL21-DE3 bacteria (transformed with PET-HCsy3). Transformation of and PET-HCsy3 plasmid into BL21-DE3 bacteria (transformed with Cd21L26). Transformation of and PET-HCsy3 plasmid into BL21-DE3 bacteria (transformed with Cd21L27). Transformation of and PET-HCsy3 plasmid into BL21-DE3 bacteria (transformed with Cd21L28). |
| 08 29 | BL21-DE3 transformants |
Growth of transformants in liquid LB. Preparing to repeat transformation. |
| 08 30 |
BL21-DE3 transformants Cd21L26 Cd21L27 Cd21L28 PET-HCsy3 |
Transformation of Cd21L26 plasmid into BL21-DE3 bacteria (transformed with PET-HCsy3). Transformation of Cd21L27 plasmid into BL21-DE3 bacteria (transformed with PET-HCsy3). Transformation of Cd21L28 plasmid into BL21-DE3 bacteria (transformed with PET-HCsy3). Transformation of and PET-HCsy3 plasmid into BL21-DE3 bacteria (transformed with Cd21L26). Transformation of and PET-HCsy3 plasmid into BL21-DE3 bacteria (transformed with Cd21L27). Transformation of and PET-HCsy3 plasmid into BL21-DE3 bacteria (transformed with Cd21L28). |
| 08 31 | BL21-DE3 transformants |
Repeating transformation for better results. Inoculating night culture of transformants. |
| 09 01 |
BL21-DE3 transformants Cd21L26 Cd21L27 Cd21L28 PET-HCsy3 |
Repeating transformation of Cd21L26 and PET-HCsy3 plasmids into BL21-DE3 bacteria, Cd21L27 and PET-HCsy3 and Cd21L28 and PET-HCsy3 into BL21-DE3 bacteria. Repeating transformation of Cd21L26, Cd21L27 and Cd21L28 plasmid into BL21-DE3 bacteria (transformed with PET-HCsy3). Repeating transformation of PET-HCsy3 plasmid into BL21-DE3 bacteria (transformed with Cd21L26, Cd21L27 and Cd21L28). Trying to improve transformation by putting less antibiotics on a Petri dish. |
| 09 02 | BL21-DE3 transformants | Checking viability of the colonies by inoculating them in liquid LB for a few hours. |
| 09 03 | BL21-DE3 transformants |
Transformants are inoculated into 20 mL LB and grown for 3 hours at 37ºC. Half of the transformants are put into 16ºC and grown overnight. Other half is centrifuged, supernatant discarded and cells put into 4ºC for further experiments. |
| 09 04 |
BL21-DE3 transformants Depleting buffer PMSF Protein Dye |
Transformants from 16ºC are centrifuged, supernatant discarded. Lysing all transformant cells with ultrasound. Lysate transferred into fresh collection tubes (1.5 mL) and used for Western Blot samples preparation. |
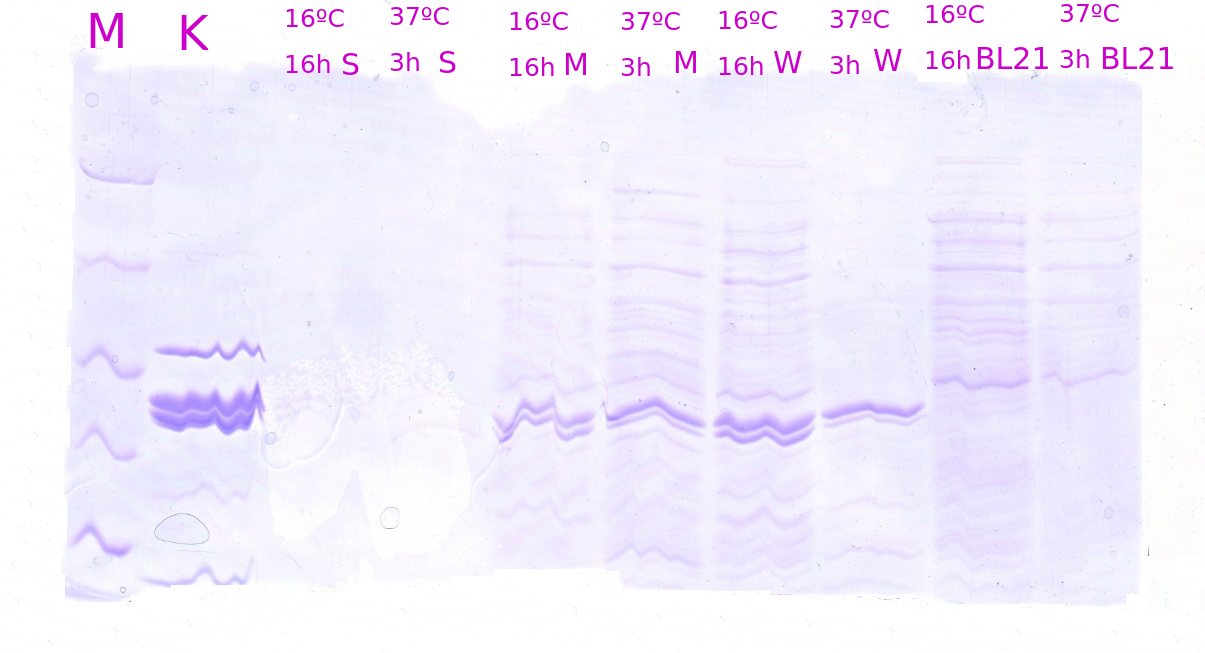
| 09 07 |
Polyacrylamid gel Protein samples |
SDS-PAGE is made in order to decide the right volume of protein solution for Western Blot. |
| 09 08 |
Polyacrylamid gel PVDF membraine Primary antibodies |
Doing Western Blot Incubating with primary antibodies overnight |
| 09 10 | Secondary antibodies |
Finishing Western Blot Incubating with secondary antibodies Visualizing blot results |
Conclusions and Results:
08 27 – Transformation unsuccessful.
08 28 – Transformation successful.
08 29 – Few colonies grown, but none multiplied in liquid LB with antibiotics.
08 31 – Few colonies have grown on a plate.
09 01 – Night culture did not grow. Transformation unsuccessful.
09 02 – Few colonies have grown, but all viable.
09 04 – Western Blot samples were made.
09 07 – Decided what volume of protein should be used for western blot.
09 10 – Visualized Western Blot results.

About
Cd21L26 - Cascade with pLux/cI right promoter and strong RBS
Cd21L27 - Cascade with pLux/cI right promoter and Medium RBS
Cd21L28 - Cascade with pLux/cI right promoter and weak RBS
| Date | Materials | Experiment |
|---|---|---|
| 09 04 |
SP1 SP3 pAcYc plasmid DH5α Karbenicilin |
Restriction of pCDF vector, SP1 and SP3 genes with EcoRI and XbaI. Ligating pieces afterwards, transforming new pCDF plasmid into bacteria. |
| 09 05 |
DH5α transformants Bl21-DE3 transformants pCola-Cas1-Cas3 plasmid Chloramphenicol Karbenicilin |
Colony PCR. Inoculating good colonies. Inoculating Bl21-DE3 transformants (BL21-DE3 with Cd21L26 + HCsy3; Cd21L27 + HCsy3; Cd21L28 + HCsy3) overnight. |
| 09 06 |
Bl21-DE3 transformants pCola-Cas1-Cas3 plasmid SP1 SP3 pCDF DH5α bacteria Chloramphenicol Kanamycin Streptomycin |
Inoculating night culture of Bl21-DE3 transformants for 3 hours. Preparing competent cells. Transforming pCola plasmid into BL21-DE3 transformants. Repeat cloning of SP1 and SP3 into pCDF vector and transforming them into DH5α. |
| 09 07 |
DH5α transformants Bl21-DE3 transformants pCola-Cas1-Cas3 plasmid Chloramphenicol Karbenicilin Kanamycin Streptomycin |
Colony PCR. Inoculating good SP1 and SP3 colonies overnight. Repeating yesterdays transformation. Inoculating night colonies again, in case of transformation failure. |
| 09 08 |
DH5α transformants Bl21-DE3 transformants pCola-Cas1-Cas3 plasmid Chloramphenicol Karbenicilin Kanamycin |
Extracting SP1 and SP3 plasmids. Repeating transformation. |
| 09 09 |
Bl21-DE3 bacteria pCola-Cas1-Cas3 plasmid Cd21L26 plasmid Cd21L27 plasmid Cd21L28 plasmid Chloramphenicol Kanamycin |
Transformation of pCola and Cd21L26 into BL21-DE3 bacteria. Transformation of pCola and Cd21L27 into BL21-DE3 bacteria. Transformation of pCola and Cd21L28 into BL21-DE3 bacteria. |
| 09 10 |
BL21-DE3 transformants Chloramphenicol Kanamycin |
Inoculating transformants overnight. |
| 09 11 |
BL21-DE3 transformants pCDF-SP1 plasmid pCDF-SP3 plasmid Chloramphenicol Kanamycin Streptomycin |
Measuring night culture optical dense. Equalizing optical dense of all bacteria. Preparing cells for transformation. Transforming pCDF-SP1, pCDF-SP3 and pCDF plasmids into BL21-DE3 transformants. |
| 09 12 |
BL21-DE3 transformants pCDF-SP1 plasmid pCDF-SP3 plasmid Chloramphenicol Kanamycin Streptomycin |
Repeating transformation. Before inoculating bacteria on a Petri dish, it is diluted 100 and 10000 times. |
| 09 13 |
BL21-DE3 transformants pCDF-SP1 plasmid pCDF-SP3 plasmid Chloramphenicol Kanamycin Streptomycin |
Repeating transformation. Before inoculating bacteria on a petri dish, it is diluted 100 and 1000 times. |
| 09 14 |
BL21-DE3 transformants pCDF-SP1 plasmid pCDF-SP3 plasmid Chloramphenicol Kanamycin Streptomycin |
Repeating transformation. Before bacteria inoculation on a petri dish, it is diluted 100 and 10000 times. |
| 09 15 |
BL21-DE3 transformants Chloramphenicol Kanamycin |
Inoculating new BL21-DE3 transformant cells. Growing overnight. |
| 09 16 |
BL21-DE3 transformants pCDF-SP1 plasmid pCDF-SP3 plasmid Chloramphenicol Kanamycin Streptomycin |
Repeating transformation. Before bacteria inoculation on a petri dish, it is diluted 100 and 10000 times. |
| 09 17 | BL21-DE3 transformants | Counting grown colonies. |
Conclusions and Results:
09 05 – Transformation successful. Colony PCR showed, that SP1 and SP3 fragments were cloned successfully.
09 06 – Mistake found. SP1 and SP3 were cloned into a wrong vector.
09 07 – Transformation was unsuccessful.
09 08 – Transformation unsuccessful. Trying to reduce antibiotic quantity.
09 09 – Transformation unsuccessful. PET-HCsy3 plasmid may have inhibited transformation.
09 10 – Transformation successful.
09 11 – BL21-DE3 transformed with Cd21L27+pCola did not grow.
09 12 – Too much bacteria grew up.
09 13 – Transformation unsuccessful. In some dishes still too many bacteria grown.
09 14 – Transformation unsuccessful.
09 15 – Transformation unsuccessful. Probably not enough pCDF-SP1 and pCDF-SP3 vector.
09 17 - Transformation successful.
About
| Date | Materials | Experiment |
|---|---|---|
| 08 26 |
P20 Carbicilin |
Transformation of P20 into JM109. |
| 08 27 |
P30 Chloramphenicol |
Transformation into JM109 (transformed with P20) |
| 08 28 |
JM109 transformants P30 Carbicilin Chloramphenicol |
Growing of JM109 transformants (low copy) and preparing for new transformation. Transformation of P30 into empty JM109. |
| 08 29 |
JM109 transformants P30 Carbicilin Chloramphenicol |
Transformation of P30 into JM109 (transformed with P20) |
| 08 30 |
JM109 transformants Carbicilin Chloramphenicol |
Transformation of P20 into JM109 (transformed with P30) |
| 08 31 |
JM109 transformants P30 P20 Carbicilin Chloramphenicol |
Transformation of P20 into JM109 (transformed with P30). Transformation of P30 into JM109 (transformed with P20). |
| 09 01 |
JM109 transformants JM109 (fresh) P20 P30 Carbicilin Chloramphenicol |
Transformation of P20 and P30 into JM109 (fresh). Transformation of P20 into JM109 (transformed with P30). Transformation of P30 into JM109 (transformed with P20). |
| 09 02 |
JM109 transformants (with P20 and P30) IPTG Carbicilin Chloramphenicol |
Growing transformants for 4 hours in a liquid LB. Transfer transformants into LB with different IPTG concentrations and grow overnight. |
| 09 12 | JM109 transformants (with P20 and P30) | Fluorescence measurement and analysis. |
| 09 06 |
JM109 transformants (with P20 and P30) IPTG Carbicilin Chloramphenicol |
Growing transformants for 4 hours in a liquid LB. Transfer transformants into LB with IPTG and grow overnight. |
| 09 07 | JM109 transformants (with P20 and P30) | Fluorescence measurement and analysis. |
| 09 08 |
JM109 transformants (with P20 and P30) IPTG Carbicilin Chloramphenicol |
Growing transformants for 4 hours in a liquid LB. Transfer transformants into LB with IPTG and grow overnight. |
| 09 10 | JM109 transformants (with P20 and P30) | Fluorescence measurement and analysis. |
| 09 11 | JM109 transformants (with P20 and P30) | Fluorescence measurement and analysis. |
| 09 13 |
JM109 transformants (with P20 and P30) pCDF vector Carbicilin Streptomycin |
Growing JM109 transformants as competent cells. Restriction and ligation of P30 fragment into pCDF vector. Transformation of new P30 into JM109 (transformed with P20). |
| 09 14 |
JM109 transformants (with P20 and P30) Carbicilin Streptomycin |
Colony PCR. |
| 09 15 |
JM109 transformants (with P20 and P30) Carbicilin Streptomycin |
Inoculating bacteria for 4 hours. Inoculating overnight with IPTG. |
| 09 16 | JM109 transformants (with P20 and P30) | Measuring fluorescence. |
| 09 17 | JM109 transformants (with P20 and P30) | Measuring fluorescence. |
Conclusions and Results:
08 26 – Transformation successful.
08 27 – Transformation unsuccessful.
08 29 – JM109 grown successfully. Transformation of P30 into empty JM109 was successful.
08 30 – Transformation unsuccessful.
08 31 - Transformation unsuccessful. The samples could have been pollen.
09 01 – Transformation unsuccessful. Maybe it was too much antibiotic on a petri dish.
09 02 – Transformation successful.
09 03 – Bad fluorimeter options.
09 07 – No repeated samples were made.
09 11 – Fluorescence measurement seems successful.
09 12 – Mistake found. P30 and P20 fragments are in an uncompetable plasmids. Repeating whole experiment.
09 14 - Transformation successful. Colony PCR successful.
09 17 - Fluorescence measurements are successful and conclusive.